SEC14L2 enables pan-genotype HCV replication in cell culture
- PMID: 26266980
- PMCID: PMC4632207
- DOI: 10.1038/nature14899
SEC14L2 enables pan-genotype HCV replication in cell culture
Abstract
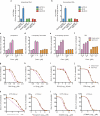
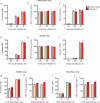
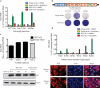
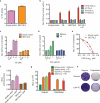
Since its discovery in 1989, efforts to grow clinical isolates of the hepatitis C virus (HCV) in cell culture have met with limited success. Only the JFH-1 isolate has the capacity to replicate efficiently in cultured hepatoma cells without cell culture-adaptive mutations. We hypothesized that cultured cells lack one or more factors required for the replication of clinical isolates. To identify the missing factors, we transduced Huh-7.5 human hepatoma cells with a pooled lentivirus-based human complementary DNA (cDNA) library, transfected the cells with HCV subgenomic replicons lacking adaptive mutations, and selected for stable replicon colonies. This led to the identification of a single cDNA, SEC14L2, that enabled RNA replication of diverse HCV genotypes in several hepatoma cell lines. This effect was dose-dependent, and required the continuous presence of SEC14L2. Full-length HCV genomes also replicated and produced low levels of infectious virus. Remarkably, SEC14L2-expressing Huh-7.5 cells also supported HCV replication following inoculation with patient sera. Mechanistic studies suggest that SEC14L2 promotes HCV infection by enhancing vitamin E-mediated protection against lipid peroxidation. This provides a foundation for development of in vitro replication systems for all HCV isolates, creating a useful platform to dissect the mechanisms by which cell culture-adaptive mutations act.
Figures






Comment in
-
Viral hepatitis: A new HCV cell culture model for the next clinical challenges.Nat Rev Gastroenterol Hepatol. 2015 Nov;12(11):611-3. doi: 10.1038/nrgastro.2015.170. Epub 2015 Oct 6. Nat Rev Gastroenterol Hepatol. 2015. PMID: 26441247
-
Pan-genotypic cell culture system for propagation of hepatitis C virus clinical isolates.Hepatology. 2016 Oct;64(4):1356-8. doi: 10.1002/hep.28751. Epub 2016 Aug 26. Hepatology. 2016. PMID: 27480678 No abstract available.
Similar articles
-
SEC14L2, a lipid-binding protein, regulates HCV replication in culture with inter- and intra-genotype variations.J Hepatol. 2019 Apr;70(4):603-614. doi: 10.1016/j.jhep.2018.11.012. Epub 2018 Nov 23. J Hepatol. 2019. PMID: 30472319
-
Viral hepatitis: A new HCV cell culture model for the next clinical challenges.Nat Rev Gastroenterol Hepatol. 2015 Nov;12(11):611-3. doi: 10.1038/nrgastro.2015.170. Epub 2015 Oct 6. Nat Rev Gastroenterol Hepatol. 2015. PMID: 26441247
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
-
Cell culture systems for the hepatitis C virus.World J Gastroenterol. 2007 May 7;13(17):2442-5. doi: 10.3748/wjg.v13.i17.2442. World J Gastroenterol. 2007. PMID: 17552027 Free PMC article. Review.
Cited by
-
FADS2-dependent fatty acid desaturation dictates cellular sensitivity to ferroptosis and permissiveness for hepatitis C virus replication.Cell Chem Biol. 2022 May 19;29(5):799-810.e4. doi: 10.1016/j.chembiol.2021.07.022. Epub 2021 Sep 13. Cell Chem Biol. 2022. PMID: 34520742 Free PMC article.
-
Hepatitis C Virus Replication.Cold Spring Harb Perspect Med. 2020 Mar 2;10(3):a037093. doi: 10.1101/cshperspect.a037093. Cold Spring Harb Perspect Med. 2020. PMID: 31570388 Free PMC article. Review.
-
Full-length sequence analysis of hepatitis C virus genotype 3b strains and development of an in vivo infectious 3b cDNA clone.J Virol. 2023 Dec 21;97(12):e0092523. doi: 10.1128/jvi.00925-23. Epub 2023 Nov 21. J Virol. 2023. PMID: 38092564 Free PMC article.
-
Hepatitis C Virus Uses Host Lipids to Its Own Advantage.Metabolites. 2021 Apr 27;11(5):273. doi: 10.3390/metabo11050273. Metabolites. 2021. PMID: 33925362 Free PMC article. Review.
-
Recent advances in understanding hepatitis C.F1000Res. 2016 Feb 3;5:F1000 Faculty Rev-131. doi: 10.12688/f1000research.7354.1. eCollection 2016. F1000Res. 2016. PMID: 26918166 Free PMC article. Review.
References
-
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. - PubMed
-
- Saeed M, et al. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology. 2013;144:56–58. e57. doi:10.1053/j.gastro.2012.09.017. - PubMed
-
- Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi:10.1002/hep.26141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

