Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo
- PMID: 26267217
- PMCID: PMC4532966
- DOI: 10.7554/eLife.07956
Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo
Abstract
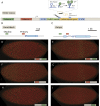
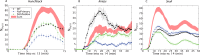
Metazoan genes are embedded in a rich milieu of regulatory information that often includes multiple enhancers possessing overlapping activities. In this study, we employ quantitative live imaging methods to assess the function of pairs of primary and shadow enhancers in the regulation of key patterning genes-knirps, hunchback, and snail-in developing Drosophila embryos. The knirps enhancers exhibit additive, sometimes even super-additive activities, consistent with classical gene fusion studies. In contrast, the hunchback enhancers function sub-additively in anterior regions containing saturating levels of the Bicoid activator, but function additively in regions where there are diminishing levels of the Bicoid gradient. Strikingly sub-additive behavior is also observed for snail, whereby removal of the proximal enhancer causes a significant increase in gene expression. Quantitative modeling of enhancer-promoter interactions suggests that weakly active enhancers function additively while strong enhancers behave sub-additively due to competition with the target promoter.
Keywords: D. melanogaster; biophysics; developmental biology; quantitative imaging; stem cells; structural biology; transcription.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

