Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- PMID: 26267277
- PMCID: PMC4534392
- DOI: 10.1371/journal.ppat.1005110
Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
Abstract
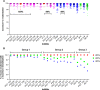
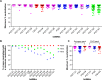
The broadly neutralizing HIV monoclonal antibodies (bnMAbs) PG9, PG16, PGT151, and PGT152 have been shown earlier to occasionally display an unusual virus neutralization profile with a non-sigmoidal slope and a plateau at <100% neutralization. In the current study, we were interested in determining the extent of non-sigmoidal slopes and plateaus at <100% for HIV bnMAbs more generally. Using both a 278 panel of pseudoviruses in a CD4 T-cell (U87.CCR5.CXCR4) assay and a panel of 117 viruses in the TZM-bl assay, we found that bnMAbs targeting many neutralizing epitopes of the spike had neutralization profiles for at least one virus that plateaued at <90%. Across both panels the bnMAbs targeting the V2 apex of Env and gp41 were most likely to show neutralization curves that plateaued <100%. Conversely, bnMAbs targeting the high-mannose patch epitopes were less likely to show such behavior. Two CD4 binding site (CD4bs) Abs also showed this behavior relatively infrequently. The phenomenon of incomplete neutralization was also observed in a large peripheral blood mononuclear cells (PBMC)-grown molecular virus clone panel derived from patient viral swarms. In addition, five bnMAbs were compared against an 18-virus panel of molecular clones produced in 293T cells and PBMCs and assayed in TZM-bl cells. Examples of plateaus <90% were seen with both types of virus production with no consistent patterns observed. In conclusion, incomplete neutralization and non-sigmoidal neutralization curves are possible for all HIV bnMAbs against a wide range of viruses produced and assayed in both cell lines and primary cells with implications for the use of antibodies in therapy and as tools for vaccine design.
Conflict of interest statement
I have read the journal's policy and the authors of the manuscript have the following competing interests: EF, HS and TW are employed by the commercial companies, Abcam Burlingame, Crucell Holland B.V. and Monogram Biosciences Inc. respectively. This does not alter our adherence to all PLOS Pathogens policies on sharing data and materials.
Figures


References
-
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, et al. (1990) Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem 265: 10373–10382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

