Expanding the ubiquitin code through post-translational modification
- PMID: 26268526
- PMCID: PMC4576978
- DOI: 10.15252/embr.201540891
Expanding the ubiquitin code through post-translational modification
Abstract
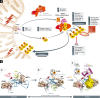
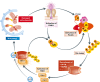
Ubiquitylation is among the most prevalent post-translational modifications (PTMs) and regulates numerous cellular functions. Interestingly, ubiquitin (Ub) can be itself modified by other PTMs, including acetylation and phosphorylation. Acetylation of Ub on K6 and K48 represses the formation and elongation of Ub chains. Phosphorylation of Ub happens on multiple sites, S57 and S65 being the most frequently modified in yeast and mammalian cells, respectively. In mammals, the PINK1 kinase activates ubiquitin ligase Parkin by phosphorylating S65 of Ub and of the Parkin Ubl domain, which in turn promotes the amplification of autophagy signals necessary for the removal of damaged mitochondria. Similarly, TBK1 phosphorylates the autophagy receptors OPTN and p62 to initiate feedback and feedforward programs for Ub-dependent removal of protein aggregates, mitochondria and pathogens (such as Salmonella and Mycobacterium tuberculosis). The impact of PINK1-mediated phosphorylation of Ub and TBK1-dependent phosphorylation of autophagy receptors (OPTN and p62) has been recently linked to the development of Parkinson's disease and amyotrophic lateral sclerosis, respectively. Hence, the post-translational modification of Ub and its receptors can efficiently expand the Ub code and modulate its functions in health and disease.
Keywords: mitophagy; phosphorylation; post‐translational modification; ubiquitin.
© 2015 The Authors.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

