Use of model organism and disease databases to support matchmaking for human disease gene discovery
- PMID: 26269093
- PMCID: PMC5473253
- DOI: 10.1002/humu.22857
Use of model organism and disease databases to support matchmaking for human disease gene discovery
Abstract
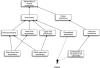
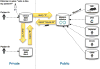
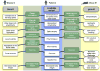
The Matchmaker Exchange application programming interface (API) allows searching a patient's genotypic or phenotypic profiles across clinical sites, for the purposes of cohort discovery and variant disease causal validation. This API can be used not only to search for matching patients, but also to match against public disease and model organism data. This public disease data enable matching known diseases and variant-phenotype associations using phenotype semantic similarity algorithms developed by the Monarch Initiative. The model data can provide additional evidence to aid diagnosis, suggest relevant models for disease mechanism and treatment exploration, and identify collaborators across the translational divide. The Monarch Initiative provides an implementation of this API for searching multiple integrated sources of data that contextualize the knowledge about any given patient or patient family into the greater biomedical knowledge landscape. While this corpus of data can aid diagnosis, it is also the beginning of research to improve understanding of rare human diseases.
Keywords: Matchmaker Exchange; informatics; model systems; ontology; phenotype; rare disease.
© 2015 WILEY PERIODICALS, INC.
Conflict of interest statement
The authors have no conflicts of interest.
Figures
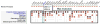
References
-
- Boycott Kym M, Vanstone Megan R, Bulman Dennis E, MacKenzie Alex E. Rare-Disease Genetics in the Era of next-Generation Sequencing: Discovery to Translation. Nature Reviews Genetics. 2013 Oct;14(10):681–91. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

