Comparison of predicted and actual consequences of missense mutations
- PMID: 26269570
- PMCID: PMC4577149
- DOI: 10.1073/pnas.1511585112
Comparison of predicted and actual consequences of missense mutations
Abstract
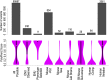
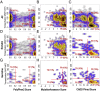
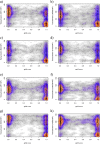
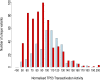
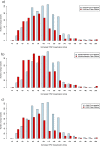
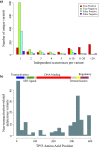
Each person's genome sequence has thousands of missense variants. Practical interpretation of their functional significance must rely on computational inferences in the absence of exhaustive experimental measurements. Here we analyzed the efficacy of these inferences in 33 de novo missense mutations revealed by sequencing in first-generation progeny of N-ethyl-N-nitrosourea-treated mice, involving 23 essential immune system genes. PolyPhen2, SIFT, MutationAssessor, Panther, CADD, and Condel were used to predict each mutation's functional importance, whereas the actual effect was measured by breeding and testing homozygotes for the expected in vivo loss-of-function phenotype. Only 20% of mutations predicted to be deleterious by PolyPhen2 (and 15% by CADD) showed a discernible phenotype in individual homozygotes. Half of all possible missense mutations in the same 23 immune genes were predicted to be deleterious, and most of these appear to become subject to purifying selection because few persist between separate mouse substrains, rodents, or primates. Because defects in immune genes could be phenotypically masked in vivo by compensation and environment, we compared inferences by the same tools with the in vitro phenotype of all 2,314 possible missense variants in TP53; 42% of mutations predicted by PolyPhen2 to be deleterious (and 45% by CADD) had little measurable consequence for TP53-promoted transcription. We conclude that for de novo or low-frequency missense mutations found by genome sequencing, half those inferred as deleterious correspond to nearly neutral mutations that have little impact on the clinical phenotype of individual cases but will nevertheless become subject to purifying selection.
Keywords: cancer; de novo mutation; evolution; immunodeficiency; nearly neutral.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Can the impact of human genetic variations be predicted?Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):11426-7. doi: 10.1073/pnas.1515057112. Epub 2015 Sep 8. Proc Natl Acad Sci U S A. 2015. PMID: 26351682 Free PMC article. No abstract available.
References
-
- Andrews TD, Sjollema G, Goodnow CC. Understanding the immunological impact of the human mutation explosion. Trends Immunol. 2013;34(3):99–106. - PubMed
-
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

