Biased Gs versus Gq proteins and β-arrestin signaling in the NK1 receptor determined by interactions in the water hydrogen bond network
- PMID: 26269596
- PMCID: PMC4591830
- DOI: 10.1074/jbc.M115.641944
Biased Gs versus Gq proteins and β-arrestin signaling in the NK1 receptor determined by interactions in the water hydrogen bond network
Abstract
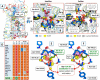
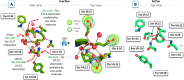
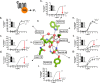
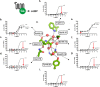
X-ray structures, molecular dynamics simulations, and mutational analysis have previously indicated that an extended water hydrogen bond network between trans-membranes I-III, VI, and VII constitutes an allosteric interface essential for stabilizing different active and inactive helical constellations during the seven-trans-membrane receptor activation. The neurokinin-1 receptor signals efficiently through Gq, Gs, and β-arrestin when stimulated by substance P, but it lacks any sign of constitutive activity. In the water hydrogen bond network the neurokinin-1 has a unique Glu residue instead of the highly conserved AspII:10 (2.50). Here, we find that this GluII:10 occupies the space of a putative allosteric modulating Na(+) ion and makes direct inter-helical interactions in particular with SerIII:15 (3.39) and AsnVII:16 (7.49) of the NPXXY motif. Mutational changes in the interface between GluII:10 and AsnVII:16 created receptors that selectively signaled through the following: 1) Gq only; 2) β-arrestin only; and 3) Gq and β-arrestin but not through Gs. Interestingly, increased constitutive Gs but not Gq signaling was observed by Ala substitution of four out of the six core polar residues of the network, in particular SerIII:15. Three residues were essential for all three signaling pathways, i.e. the water-gating micro-switch residues TrpVI:13 (6.48) of the CWXP motif and TyrVII:20 (7.53) of the NPXXY motif plus the totally conserved AsnI:18 (1.50) stabilizing the kink in trans-membrane VII. It is concluded that the interface between position II:10 (2.50), III:15 (3.39), and VII:16 (7.49) in the center of the water hydrogen bond network constitutes a focal point for fine-tuning seven trans-membrane receptor conformations activating different signal transduction pathways.
Keywords: G protein-coupled receptor (GPCR); cell signaling; functional selectivity; homology modeling; hydrogen bond network; molecular pharmacology; receptor structure-function.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Nygaard R., Frimurer T. M., Holst B., Rosenkilde M. M., Schwartz T. W. (2009) Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 30, 249–259 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

