Role of the normal gut microbiota
- PMID: 26269668
- PMCID: PMC4528021
- DOI: 10.3748/wjg.v21.i29.8787
Role of the normal gut microbiota
Abstract
Relation between the gut microbiota and human health is being increasingly recognised. It is now well established that a healthy gut flora is largely responsible for overall health of the host. The normal human gut microbiota comprises of two major phyla, namely Bacteroidetes and Firmicutes. Though the gut microbiota in an infant appears haphazard, it starts resembling the adult flora by the age of 3 years. Nevertheless, there exist temporal and spatial variations in the microbial distribution from esophagus to the rectum all along the individual's life span. Developments in genome sequencing technologies and bioinformatics have now enabled scientists to study these microorganisms and their function and microbe-host interactions in an elaborate manner both in health and disease. The normal gut microbiota imparts specific function in host nutrient metabolism, xenobiotic and drug metabolism, maintenance of structural integrity of the gut mucosal barrier, immunomodulation, and protection against pathogens. Several factors play a role in shaping the normal gut microbiota. They include (1) the mode of delivery (vaginal or caesarean); (2) diet during infancy (breast milk or formula feeds) and adulthood (vegan based or meat based); and (3) use of antibiotics or antibiotic like molecules that are derived from the environment or the gut commensal community. A major concern of antibiotic use is the long-term alteration of the normal healthy gut microbiota and horizontal transfer of resistance genes that could result in reservoir of organisms with a multidrug resistant gene pool.
Keywords: Bioinformatics; Health; Immunomodulation; Metabolic function; Normal gut microbiota.
Figures
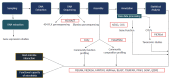
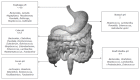

References
-
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. - PubMed
-
- Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Müller G, Stokholm J, Smith B, Krogfelt KA. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–652.e1-5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

