Effect of IKZF1 deletions on signal transduction pathways in Philadelphia chromosome negative pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL)
- PMID: 26269779
- PMCID: PMC4534008
- DOI: 10.1186/s40164-015-0017-y
Effect of IKZF1 deletions on signal transduction pathways in Philadelphia chromosome negative pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL)
Abstract
Background: IKZF1 deletions are an unfavorable prognostic factor in children with Philadelphia chromosome positive (Ph(+)) as well as negative (Ph(-)) acute lymphoblastic leukemia (ALL). Although IKZF1 deletions occur in 10-15% of Ph(-) ALL cases, effects of IKZF1 deletions on signaling pathways in this group have not been extensively studied. Therefore, in this study we aimed to study the effect of IKZF1 deletions on active signal transduction pathways.
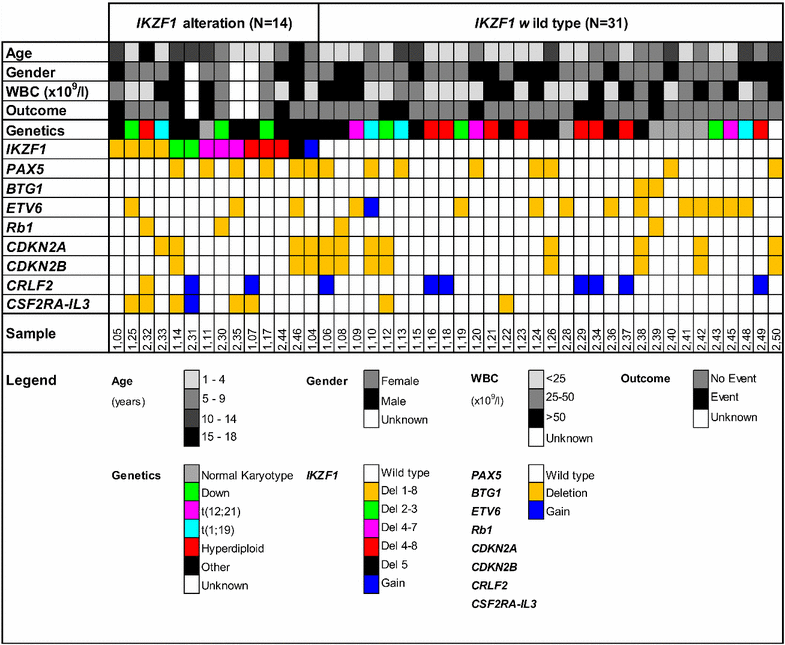
Methods: Multiplex ligation-dependent probe amplification (MLPA) was used to determine IKZF1 deletions and other copy number alterations in 109 pediatric B-Cell Precursor ALL (BCP-ALL) patients. Kinase activity profiling of 45 primary Ph(-) BCP-ALL patients (31 IKZF1 wild type patients and 14 patients harboring an IKZF1 alteration) and western blot analysis of 14 pediatric BCP-ALL samples was performed to determine active signal transduction pathways.
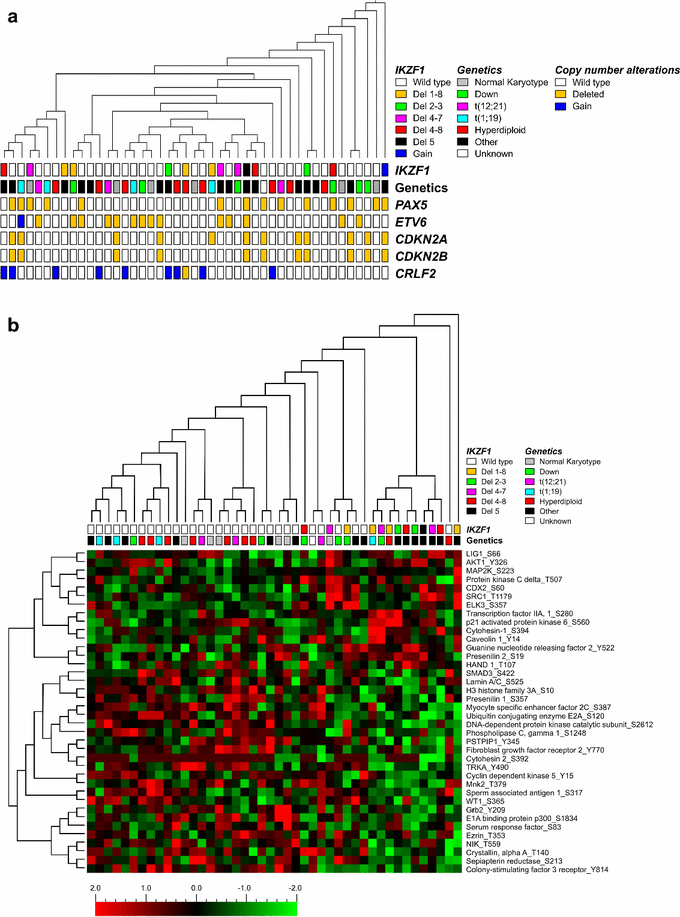
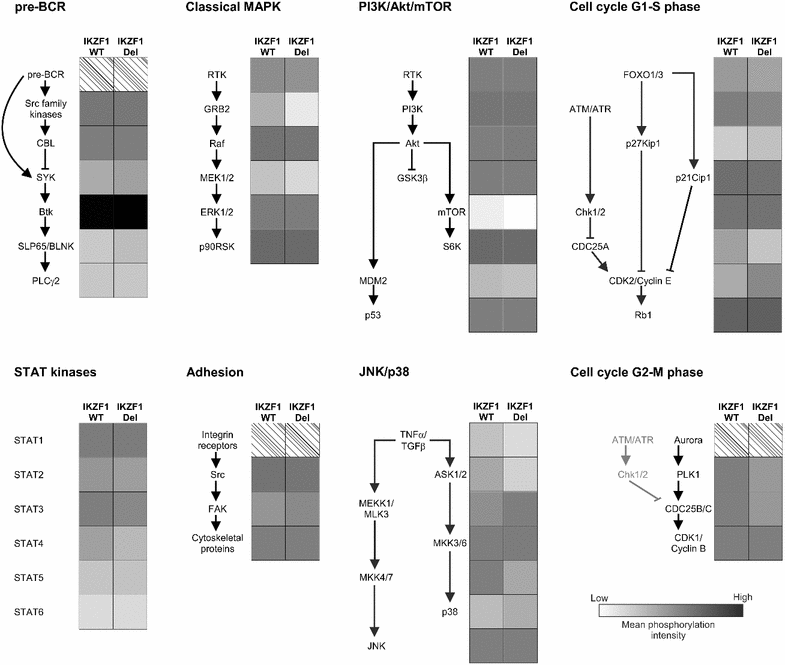
Results: Unsupervised hierarchical cluster analysis of kinome profiles of 45 pediatric Ph(-) ALL cases showed no clustering based on IKZF1 status. Comparing the phosphorylation intensities of peptides associated with signaling pathways known to be involved in BCP-ALL maintenance, we did not observe differences between the two groups. Western blot analysis of 14 pediatric BCP-ALL samples showed large variations in phosphorylation levels between the different ALL samples, independent of IKZF1 status.
Conclusions: Based on these results we conclude that, although IKZF1 deletions appear to be an important clinical prognostic factor, we were unable to identify a unique IKZF1 dependent protein expression signature in pediatric Ph(-) ALL and consequently no specific targets for future therapy of Ph(-) IKZF1 deleted BCP-ALL could be identified.
Keywords: Acute lymphoblastic leukemia; IKZF1; Kinome profiling; Signaling.
Figures

References
-
- van der Veer A, Waanders E, Pieters R, Willemse ME, Van Reijmersdal SV, Russell LJ, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–2629. doi: 10.1182/blood-2012-10-462358. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

