Distribution, Numbers, and Diversity of ESBL-Producing E. coli in the Poultry Farm Environment
- PMID: 26270644
- PMCID: PMC4536194
- DOI: 10.1371/journal.pone.0135402
Distribution, Numbers, and Diversity of ESBL-Producing E. coli in the Poultry Farm Environment
Abstract
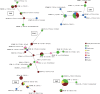
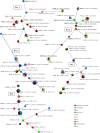
This study aimed to discern the contribution of poultry farms to the contamination of the environment with ESBL-producing Escherichia coli and therewith, potentially to the spread of these bacteria to humans and other animals. ESBL-producing E. coli were detected at all investigated laying hen farms (n = 5) and broiler farms (n = 3) in 65% (46/71) and 81% (57/70) of poultry faeces samples, respectively. They were detected in rinse water and run-off water (21/26; 81%), other farm animals (11/14; 79%), dust (21/35; 60%), surface water adjacent to farms (20/35; 57%), soil (48/87; 55%), on flies (11/73; 15%), and in barn air (2/33; 6%). The highest prevalence and concentrations in the outdoor environment were observed in soil of free-range areas at laying hen farms (100% of samples positive, geometric mean concentration 2.4×10(4) cfu/kg), and surface waters adjacent to broiler farms during, or shortly after, cleaning between production rounds (91% of samples positive, geometric mean concentration 1.9×10(2) cfu/l). The diversity of ESBL-producing E. coli variants with respect to sequence type, phylogenetic group, ESBL-genotype and antibiotic resistance profile was high, especially on broiler farms where on average 16 different variants were detected, and the average Simpson's Indices of diversity (SID; 1-D) were 0.93 and 0.94 among flock and environmental isolates respectively. At laying hen farms on average nine variants were detected, with SIDs of 0.63 (flock isolates) and 0.77 (environmental isolates). Sixty percent of environmental isolates were identical to flock isolates at the same farm. The highest proportions of 'flock variants' were observed in dust (94%), run-off gullies (82%), and barn air (67%), followed by surface water (57%), soil (56%), flies (50%) and other farm animals (35%).The introduction of ESBL-producing E. coli from poultry farms to the environment may pose a health risk if these bacteria reach places where people may become exposed.
Conflict of interest statement
Figures




References
-
- Kümmerer K. Resistance in the environment. J Antimicrob Chemother. 2004;54: 311–320. - PubMed
-
- Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, et al. Prevalence and spread of extended-spectrum ß-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14 Suppl. 1: 144–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

