Allosteric Activation of Bacterial Swi2/Snf2 (Switch/Sucrose Non-fermentable) Protein RapA by RNA Polymerase: BIOCHEMICAL AND STRUCTURAL STUDIES
- PMID: 26272746
- PMCID: PMC4583045
- DOI: 10.1074/jbc.M114.618801
Allosteric Activation of Bacterial Swi2/Snf2 (Switch/Sucrose Non-fermentable) Protein RapA by RNA Polymerase: BIOCHEMICAL AND STRUCTURAL STUDIES
Abstract
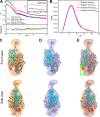
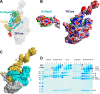
Members of the Swi2/Snf2 (switch/sucrose non-fermentable) family depend on their ATPase activity to mobilize nucleic acid-protein complexes for gene expression. In bacteria, RapA is an RNA polymerase (RNAP)-associated Swi2/Snf2 protein that mediates RNAP recycling during transcription. It is known that the ATPase activity of RapA is stimulated by its interaction with RNAP. It is not known, however, how the RapA-RNAP interaction activates the enzyme. Previously, we determined the crystal structure of RapA. The structure revealed the dynamic nature of its N-terminal domain (Ntd), which prompted us to elucidate the solution structure and activity of both the full-length protein and its Ntd-truncated mutant (RapAΔN). Here, we report the ATPase activity of RapA and RapAΔN in the absence or presence of RNAP and the solution structures of RapA and RapAΔN either ligand-free or in complex with RNAP. Determined by small-angle x-ray scattering, the solution structures reveal a new conformation of RapA, define the binding mode and binding site of RapA on RNAP, and show that the binding sites of RapA and σ(70) on the surface of RNAP largely overlap. We conclude that the ATPase activity of RapA is inhibited by its Ntd but stimulated by RNAP in an allosteric fashion and that the conformational changes of RapA and its interaction with RNAP are essential for RNAP recycling. These and previous findings outline the functional cycle of RapA, which increases our understanding of the mechanism and regulation of Swi2/Snf2 proteins in general and of RapA in particular. The new structural information also leads to a hypothetical model of RapA in complex with RNAP immobilized during transcription.
Keywords: ATPase; RNA polymerase; RNA polymerase recycling; RapA-RNA polymerase interaction; allosteric activation; bacterial Swi2/Snf2 protein RapA; bacterial transcription; small-angle x-ray scattering (SAXS); transcription factor.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Muzzin O., Campbell E. A., Xia L., Severinova E., Darst S. A., Severinov K. (1998) Disruption of Escherichia coli hepA, an RNA polymerase-associated protein, causes UV sensitivity. J. Biol. Chem. 273, 15157–15161 - PubMed
-
- Sukhodolets M. V., Jin D. J. (1998) RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 273, 7018–7023 - PubMed
-
- Sukhodolets M. V., Jin D. J. (2000) Interaction between RNA polymerase and RapA, a bacterial homolog of the SWI/SNF protein family. J. Biol. Chem. 275, 22090–22097 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

