Complete biosynthesis of opioids in yeast
- PMID: 26272907
- PMCID: PMC4924617
- DOI: 10.1126/science.aac9373
Complete biosynthesis of opioids in yeast
Abstract
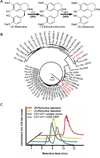
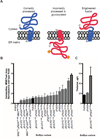
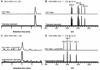
Opioids are the primary drugs used in Western medicine for pain management and palliative care. Farming of opium poppies remains the sole source of these essential medicines, despite diverse market demands and uncertainty in crop yields due to weather, climate change, and pests. We engineered yeast to produce the selected opioid compounds thebaine and hydrocodone starting from sugar. All work was conducted in a laboratory that is permitted and secured for work with controlled substances. We combined enzyme discovery, enzyme engineering, and pathway and strain optimization to realize full opiate biosynthesis in yeast. The resulting opioid biosynthesis strains required the expression of 21 (thebaine) and 23 (hydrocodone) enzyme activities from plants, mammals, bacteria, and yeast itself. This is a proof of principle, and major hurdles remain before optimization and scale-up could be achieved. Open discussions of options for governing this technology are also needed in order to responsibly realize alternative supplies for these medically relevant compounds.
Copyright © 2015, American Association for the Advancement of Science.
Figures

Comment in
-
BIOENGINEERING. Yeast cell factories on the horizon.Science. 2015 Sep 4;349(6252):1050-1. doi: 10.1126/science.aad2081. Science. 2015. PMID: 26339012 No abstract available.
-
Biological synthesis unbounded?Nat Biotechnol. 2015 Nov;33(11):1148-9. doi: 10.1038/nbt.3399. Nat Biotechnol. 2015. PMID: 26544145 No abstract available.
References
-
- World Health Organization. Geneva, Switzerland: 2013. 18th WHO essential medicines list.
-
- Seya MJ, Gelders SF, Achara OU, Milani B, Scholten WK. A first comparison between the consumption of and the need for opioid analgesics at country, regional, and global levels. J Pain Palliat. Care Pharmacother. 2011;25:6–18. - PubMed
-
- International Narcotics Control Board (INCB) Narcotic drugs: Estimated world requirements for 2015 - Statistics for 2013. 2014
-
- Bradsher K. Shake-up on opium island. New York Times. 2014
-
- Reed JW, Hudlicky T. The quest for a practical synthesis of morphine alkaloids and their derivatives by chemoenzymatic methods. Acc. Chem. Res. 2015;48:674–687. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

