OVA: integrating molecular and physical phenotype data from multiple biomedical domain ontologies with variant filtering for enhanced variant prioritization
- PMID: 26272982
- PMCID: PMC4653395
- DOI: 10.1093/bioinformatics/btv473
OVA: integrating molecular and physical phenotype data from multiple biomedical domain ontologies with variant filtering for enhanced variant prioritization
Abstract
Motivation: Exome sequencing has become a de facto standard method for Mendelian disease gene discovery in recent years, yet identifying disease-causing mutations among thousands of candidate variants remains a non-trivial task.
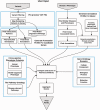
Results: Here we describe a new variant prioritization tool, OVA (ontology variant analysis), in which user-provided phenotypic information is exploited to infer deeper biological context. OVA combines a knowledge-based approach with a variant-filtering framework. It reduces the number of candidate variants by considering genotype and predicted effect on protein sequence, and scores the remainder on biological relevance to the query phenotype.We take advantage of several ontologies in order to bridge knowledge across multiple biomedical domains and facilitate computational analysis of annotations pertaining to genes, diseases, phenotypes, tissues and pathways. In this way, OVA combines information regarding molecular and physical phenotypes and integrates both human and model organism data to effectively prioritize variants. By assessing performance on both known and novel disease mutations, we show that OVA performs biologically meaningful candidate variant prioritization and can be more accurate than another recently published candidate variant prioritization tool.
Availability and implementation: OVA is freely accessible at http://dna2.leeds.ac.uk:8080/OVA/index.jsp.
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: umaan@leeds.ac.uk.
© The Author 2015. Published by Oxford University Press.
Figures


References
-
- Adie E.A., et al. (2006) SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics, 22, 773–774. - PubMed
-
- Armstrong R.A. (2014) When to use the Bonferroni correction. Ophthalmic Physiol. Opt., 34, 502–508. - PubMed
-
- Bornigen D., et al. (2012) An unbiased evaluation of gene prioritization tools. Bioinformatics, 28, 3081–3088. - PubMed

