Prediction of MicroRNA-Disease Associations Based on Social Network Analysis Methods
- PMID: 26273645
- PMCID: PMC4529919
- DOI: 10.1155/2015/810514
Prediction of MicroRNA-Disease Associations Based on Social Network Analysis Methods
Abstract
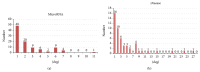
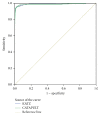
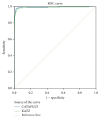
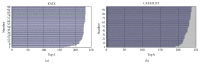
MicroRNAs constitute an important class of noncoding, single-stranded, ~22 nucleotide long RNA molecules encoded by endogenous genes. They play an important role in regulating gene transcription and the regulation of normal development. MicroRNAs can be associated with disease; however, only a few microRNA-disease associations have been confirmed by traditional experimental approaches. We introduce two methods to predict microRNA-disease association. The first method, KATZ, focuses on integrating the social network analysis method with machine learning and is based on networks derived from known microRNA-disease associations, disease-disease associations, and microRNA-microRNA associations. The other method, CATAPULT, is a supervised machine learning method. We applied the two methods to 242 known microRNA-disease associations and evaluated their performance using leave-one-out cross-validation and 3-fold cross-validation. Experiments proved that our methods outperformed the state-of-the-art methods.
Figures
References
-
- Jiménez P., Thomas F., Torras C. 3D collision detection: A survey. Computers & Graphics. 2001;25(2):269–285. doi: 10.1016/s0097-8493(00)00130-8. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

