Context Sensitive Modeling of Cancer Drug Sensitivity
- PMID: 26274927
- PMCID: PMC4537214
- DOI: 10.1371/journal.pone.0133850
Context Sensitive Modeling of Cancer Drug Sensitivity
Abstract
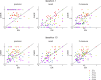
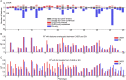
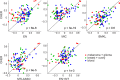
Recent screening of drug sensitivity in large panels of cancer cell lines provides a valuable resource towards developing algorithms that predict drug response. Since more samples provide increased statistical power, most approaches to prediction of drug sensitivity pool multiple cancer types together without distinction. However, pan-cancer results can be misleading due to the confounding effects of tissues or cancer subtypes. On the other hand, independent analysis for each cancer-type is hampered by small sample size. To balance this trade-off, we present CHER (Contextual Heterogeneity Enabled Regression), an algorithm that builds predictive models for drug sensitivity by selecting predictive genomic features and deciding which ones should-and should not-be shared across different cancers, tissues and drugs. CHER provides significantly more accurate models of drug sensitivity than comparable elastic-net-based models. Moreover, CHER provides better insight into the underlying biological processes by finding a sparse set of shared and type-specific genomic features.
Conflict of interest statement
Figures



Similar articles
-
Prediction of Chemosensitivity in Multiple Primary Cancer Patients Using Machine Learning.Anticancer Res. 2021 May;41(5):2419-2429. doi: 10.21873/anticanres.15017. Anticancer Res. 2021. PMID: 33952467
-
Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data.Pac Symp Biocomput. 2014:63-74. Pac Symp Biocomput. 2014. PMID: 24297534 Free PMC article.
-
DISIS: prediction of drug response through an iterative sure independence screening.PLoS One. 2015 Mar 20;10(3):e0120408. doi: 10.1371/journal.pone.0120408. eCollection 2015. PLoS One. 2015. PMID: 25794193 Free PMC article.
-
Cell line modeling for systems medicine in cancers (review).Int J Oncol. 2014 Feb;44(2):371-6. doi: 10.3892/ijo.2013.2202. Epub 2013 Dec 2. Int J Oncol. 2014. PMID: 24297677 Free PMC article. Review.
-
Genomic approach towards personalized anticancer drug therapy.Pharmacogenomics. 2012 Jan;13(2):191-9. doi: 10.2217/pgs.11.157. Pharmacogenomics. 2012. PMID: 22256868 Review.
Cited by
-
Tissue specificity of in vitro drug sensitivity.J Am Med Inform Assoc. 2018 Feb 1;25(2):158-166. doi: 10.1093/jamia/ocx062. J Am Med Inform Assoc. 2018. PMID: 29016819 Free PMC article.
-
Systematic identification of feature combinations for predicting drug response with Bayesian multi-view multi-task linear regression.Bioinformatics. 2017 Jul 15;33(14):i359-i368. doi: 10.1093/bioinformatics/btx266. Bioinformatics. 2017. PMID: 28881998 Free PMC article.
-
Impact of between-tissue differences on pan-cancer predictions of drug sensitivity.PLoS Comput Biol. 2021 Feb 25;17(2):e1008720. doi: 10.1371/journal.pcbi.1008720. eCollection 2021 Feb. PLoS Comput Biol. 2021. PMID: 33630864 Free PMC article.
-
Computational models for predicting drug responses in cancer research.Brief Bioinform. 2017 Sep 1;18(5):820-829. doi: 10.1093/bib/bbw065. Brief Bioinform. 2017. PMID: 27444372 Free PMC article. Review.
-
Context-specific functional module based drug efficacy prediction.BMC Bioinformatics. 2016 Jul 28;17 Suppl 6(Suppl 6):275. doi: 10.1186/s12859-016-1078-6. BMC Bioinformatics. 2016. PMID: 27490093 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

