RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself
- PMID: 26275108
- PMCID: PMC5444807
- DOI: 10.1126/science.aac7049
RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself
Abstract
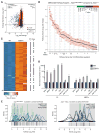
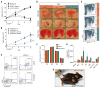
Adenosine-to-inosine (A-to-I) editing is a highly prevalent posttranscriptional modification of RNA, mediated by ADAR (adenosine deaminase acting on RNA) enzymes. In addition to RNA editing, additional functions have been proposed for ADAR1. To determine the specific role of RNA editing by ADAR1, we generated mice with an editing-deficient knock-in mutation (Adar1(E861A), where E861A denotes Glu(861)→Ala(861)). Adar1(E861A/E861A) embryos died at ~E13.5 (embryonic day 13.5), with activated interferon and double-stranded RNA (dsRNA)-sensing pathways. Genome-wide analysis of the in vivo substrates of ADAR1 identified clustered hyperediting within long dsRNA stem loops within 3' untranslated regions of endogenous transcripts. Finally, embryonic death and phenotypes of Adar1(E861A/E861A) were rescued by concurrent deletion of the cytosolic sensor of dsRNA, MDA5. A-to-I editing of endogenous dsRNA is the essential function of ADAR1, preventing the activation of the cytosolic dsRNA response by endogenous transcripts.
Copyright © 2015, American Association for the Advancement of Science.
Figures


Comment in
-
RNA editing by ADAR1 marks dsRNA as "self".Cell Res. 2015 Dec;25(12):1283-4. doi: 10.1038/cr.2015.135. Epub 2015 Nov 24. Cell Res. 2015. PMID: 26596786 Free PMC article.
-
A-to-I editing prevents self-RNA sensing.Nat Rev Mol Cell Biol. 2023 Feb;24(2):85. doi: 10.1038/s41580-022-00540-4. Epub 2022 Sep 8. Nat Rev Mol Cell Biol. 2023. PMID: 36076034 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

