Impact of the Regulators SigB, Rot, SarA and sarS on the Toxic Shock Tst Promoter and TSST-1 Expression in Staphylococcus aureus
- PMID: 26275216
- PMCID: PMC4537247
- DOI: 10.1371/journal.pone.0135579
Impact of the Regulators SigB, Rot, SarA and sarS on the Toxic Shock Tst Promoter and TSST-1 Expression in Staphylococcus aureus
Abstract
Staphylococcus aureus is an important pathogen manifesting virulence through diverse disease forms, ranging from acute skin infections to life-threatening bacteremia or systemic toxic shock syndromes. In the latter case, the prototypical superantigen is TSST-1 (Toxic Shock Syndrome Toxin 1), encoded by tst(H), and carried on a mobile genetic element that is not present in all S. aureus strains. Transcriptional regulation of tst is only partially understood. In this study, we dissected the role of sarA, sarS (sarH1), RNAIII, rot, and the alternative stress sigma factor sigB (σB). By examining tst promoter regulation predominantly in the context of its native sequence within the SaPI1 pathogenicity island of strain RN4282, we discovered that σB emerged as a particularly important tst regulator. We did not detect a consensus σB site within the tst promoter, and thus the effect of σB is likely indirect. We found that σB strongly repressed the expression of the toxin via at least two distinct regulatory pathways dependent upon sarA and agr. Furthermore rot, a member of SarA family, was shown to repress tst expression when overexpressed, although its deletion had no consistent measurable effect. We could not find any detectable effect of sarS, either by deletion or overexpression, suggesting that this regulator plays a minimal role in TSST-1 expression except when combined with disruption of sarA. Collectively, our results extend our understanding of complex multifactorial regulation of tst, revealing several layers of negative regulation. In addition to environmental stimuli thought to impact TSST-1 production, these findings support a model whereby sporadic mutation in a few key negative regulators can profoundly affect and enhance TSST-1 expression.
Conflict of interest statement
Figures
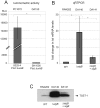
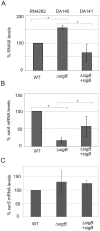




References
-
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–32. Epub 1998/08/26. . - PubMed
-
- Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49(2):93–105. Epub 2003/05/03. doi: S0147619X02001579 [pii]. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

