GC skew defines distinct RNA polymerase pause sites in CpG island promoters
- PMID: 26275623
- PMCID: PMC4617957
- DOI: 10.1101/gr.189068.114
GC skew defines distinct RNA polymerase pause sites in CpG island promoters
Abstract
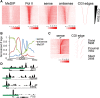
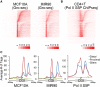
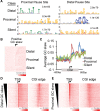
CpG islands (CGIs) are associated with over half of human gene promoters and are characterized by a unique chromatin environment and high levels of bidirectional transcriptional activity relative to surrounding genomic regions, suggesting that RNA polymerase (Pol II) progression past the CGI boundaries is restricted. Here we describe a novel transcriptional regulatory step wherein Pol II encounters an additional barrier to elongation distinct from the promoter-proximal pause and occurring at the downstream boundary of the CGI domain. For most CGI-associated promoters, Pol II exhibits a dominant pause at either the promoter-proximal or this distal site that correlates, both in position and in intensity, with local regions of high GC skew, a sequence feature known to form unique secondary structures. Upon signal-induced gene activation, long-range enhancer contacts at the dominant pause site are selectively enhanced, suggesting a new role for enhancers at the downstream pause. These data point to an additional level of control over transcriptional output at a subset of CGI-associated genes that is linked to DNA sequence and the integrity of the CGI domain.
© 2015 Kellner et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
