NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins
- PMID: 26275778
- PMCID: PMC4605303
- DOI: 10.1093/nar/gkv809
NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins
Abstract
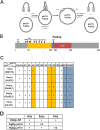
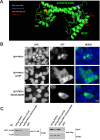
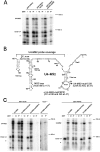
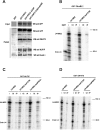
The Sm proteins are loaded on snRNAs by the SMN complex, but how snRNP-specific proteins are assembled remains poorly characterized. U4 snRNP and box C/D snoRNPs have structural similarities. They both contain the 15.5K and proteins with NOP domains (PRP31 for U4, NOP56/58 for snoRNPs). Biogenesis of box C/D snoRNPs involves NUFIP and the HSP90/R2TP chaperone system and here, we explore the function of this machinery in U4 RNP assembly. We show that yeast Prp31 interacts with several components of the NUFIP/R2TP machinery, and that these interactions are separable from each other. In human cells, PRP31 mutants that fail to stably associate with U4 snRNA still interact with components of the NUFIP/R2TP system, indicating that these interactions precede binding of PRP31 to U4 snRNA. Knock-down of NUFIP leads to mislocalization of PRP31 and decreased association with U4. Moreover, NUFIP is associated with the SMN complex through direct interactions with Gemin3 and Gemin6. Altogether, our data suggest a model in which the NUFIP/R2TP system is connected with the SMN complex and facilitates assembly of U4 snRNP-specific proteins.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- van der Feltz C., Anthony K., Brilot A., Pomeranz Krummel D. Architecture of the spliceosome. Biochemistry. 2012;51:3321–3333. - PubMed
-
- Wahl M., Will C., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Dunn E., Rader S. Secondary structure of U6 small nuclear RNA: implications for spliceosome assembly. Biochem. Soc. Trans. 2010;38:1099–1104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

