Comparative genome analysis of Mycoplasma pneumoniae
- PMID: 26275904
- PMCID: PMC4537597
- DOI: 10.1186/s12864-015-1801-0
Comparative genome analysis of Mycoplasma pneumoniae
Abstract
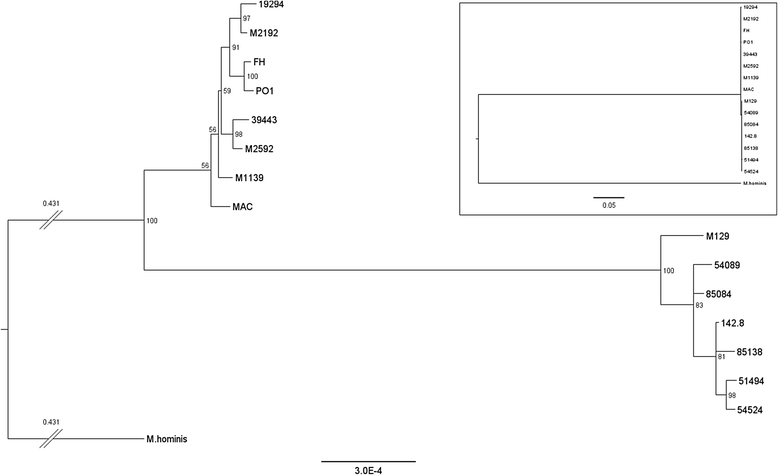
Background: Mycoplasma pneumoniae is a common pathogen that causes upper and lower respiratory tract infections in people of all ages, responsible for up to 40% of community-acquired pneumonias. It also causes a wide array of extrapulmonary infections and autoimmune phenomena. Phylogenetic studies of the organism have been generally restricted to specific genes or regions of the genome, because whole genome sequencing has been completed for only 4 strains. To better understand the physiology and pathogenicity of this important human pathogen, we performed comparative genomic analysis of 15 strains of M. pneumoniae that were isolated between the 1940s to 2009 from respiratory specimens and cerebrospinal fluid originating from the USA, China and England.
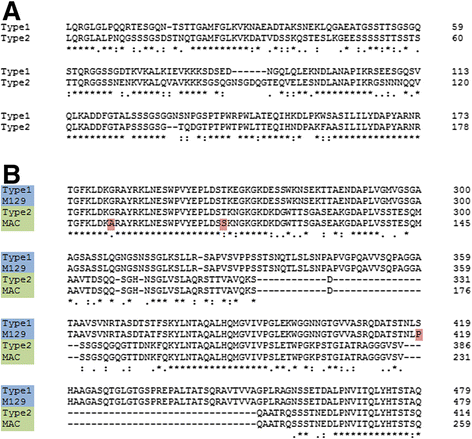
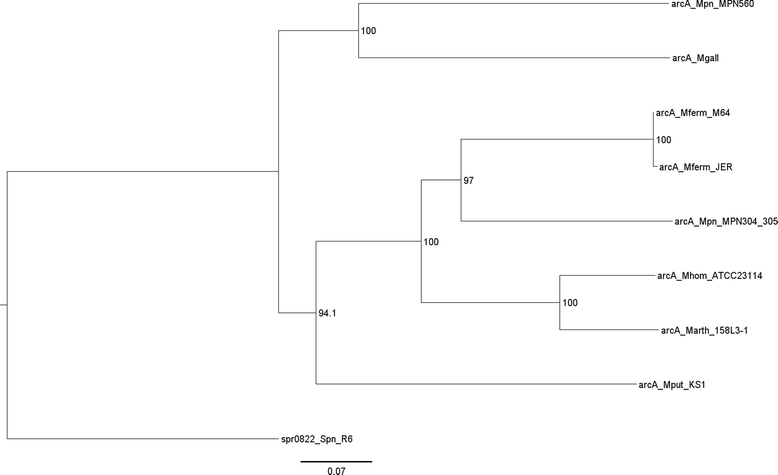
Results: Illumina MiSeq whole genome sequencing was performed on the 15 strains and all genome sequences were completed. Results from the comparative genomic analysis indicate that although about 1500 SNP and indel variants exist between type1 and type 2 strains, there is an overall high degree of sequence similarity among the strains (>99% identical to each other). Within the two subtypes, conservation of most genes, including the CARDS toxin gene and arginine deiminase genes, was observed. The major variation occurs in the P1 and ORF6 genes associated with the adhesin complex. Multiple hsdS genes (encodes S subunit of type I restriction enzyme) with variable tandem repeat copy numbers were found in all 15 genomes.
Conclusions: These data indicate that despite conclusions drawn from 16S rRNA sequences suggesting rapid evolution, the M. pneumoniae genome is extraordinarily stable over time and geographic distance across the globe with a striking lack of evidence of horizontal gene transfer.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

