Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma
- PMID: 26276629
- PMCID: PMC4538708
- DOI: 10.1016/j.cell.2015.07.019
Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma
Abstract
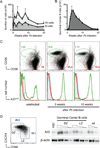
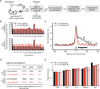
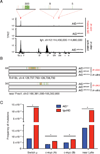
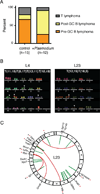
Chronic infection with Plasmodium falciparum was epidemiologically associated with endemic Burkitt's lymphoma, a mature B cell cancer characterized by chromosome translocation between the c-myc oncogene and Igh, over 50 years ago. Whether infection promotes B cell lymphoma, and if so by which mechanism, remains unknown. To investigate the relationship between parasitic disease and lymphomagenesis, we used Plasmodium chabaudi (Pc) to produce chronic malaria infection in mice. Pc induces prolonged expansion of germinal centers (GCs), unique compartments in which B cells undergo rapid clonal expansion and express activation-induced cytidine deaminase (AID), a DNA mutator. GC B cells elicited during Pc infection suffer widespread DNA damage, leading to chromosome translocations. Although infection does not change the overall rate, it modifies lymphomagenesis to favor mature B cell lymphomas that are AID dependent and show chromosome translocations. Thus, malaria infection favors mature B cell cancers by eliciting protracted AID expression in GC B cells. PAPERCLIP.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Tumour immunology: Malaria alters B cell lymphomagenesis.Nat Rev Immunol. 2015 Sep 15;15(9):528. doi: 10.1038/nri3906. Epub 2015 Aug 21. Nat Rev Immunol. 2015. PMID: 26292639 No abstract available.
References
-
- Achtman AH, Stephens R, Cadman ET, Harrison V, Langhorne J. Malaria-specific antibody responses and parasite persistence after infection of mice with Plasmodium chabaudi chabaudi. Parasite immunology. 2007;29:435–444. - PubMed
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

