Comparative Genomics of Environmental and Clinical Stenotrophomonas maltophilia Strains with Different Antibiotic Resistance Profiles
- PMID: 26276674
- PMCID: PMC4607518
- DOI: 10.1093/gbe/evv161
Comparative Genomics of Environmental and Clinical Stenotrophomonas maltophilia Strains with Different Antibiotic Resistance Profiles
Abstract
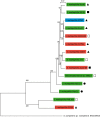
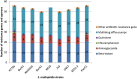
Stenotrophomonas maltophilia, a ubiquitous Gram-negative γ-proteobacterium, has emerged as an important opportunistic pathogen responsible for nosocomial infections. A major characteristic of clinical isolates is their high intrinsic or acquired antibiotic resistance level. The aim of this study was to decipher the genetic determinism of antibiotic resistance among strains from different origins (i.e., natural environment and clinical origin) showing various antibiotic resistance profiles. To this purpose, we selected three strains isolated from soil collected in France or Burkina Faso that showed contrasting antibiotic resistance profiles. After whole-genome sequencing, the phylogenetic relationships of these 3 strains and 11 strains with available genome sequences were determined. Results showed that a strain's phylogeny did not match their origin or antibiotic resistance profiles. Numerous antibiotic resistance coding genes and efflux pump operons were revealed by the genome analysis, with 57% of the identified genes not previously described. No major variation in the antibiotic resistance gene content was observed between strains irrespective of their origin and antibiotic resistance profiles. Although environmental strains generally carry as many multidrug resistant (MDR) efflux pumps as clinical strains, the absence of resistance-nodulation-division (RND) pumps (i.e., SmeABC) previously described to be specific to S. maltophilia was revealed in two environmental strains (BurA1 and PierC1). Furthermore the genome analysis of the environmental MDR strain BurA1 showed the absence of SmeABC but the presence of another putative MDR RND efflux pump, named EbyCAB on a genomic island probably acquired through horizontal gene transfer.
Keywords: Stenotrophomonas; antibiotic resistance; efflux pump; phylogeny.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Akova M, Bonfiglio G, Livermore DM. 1991. Susceptibility to beta-lactam antibiotics of mutant strains of Xanthomonas maltophilia with high-level and low-level constitutive expression of L1 and L2 beta-lactamases. J Med Microbiol. 35:208–213. - PubMed
-
- Al-Hamad A, Upton M, Burnie J. 2009. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J Antimicrob Chemother. 64:731–734. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

