Amino acid management in cancer
- PMID: 26277542
- PMCID: PMC4800996
- DOI: 10.1016/j.semcdb.2015.08.002
Amino acid management in cancer
Abstract
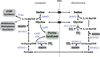
Amino acids have a dual role in cellular metabolism, as they are both the building blocks for protein synthesis and intermediate metabolites which fuel other biosynthetic reactions. Recent work has demonstrated that deregulation of both arms of amino acid management are common alterations seen in cancer. Among the most highly consumed nutrients by cancer cells are the amino acids glutamine and serine, and the biosynthetic pathways that metabolize them are required in various cancer subtypes and the object of current efforts to target cancer metabolism. Also altered in cancer are components of the machinery which sense amino acid sufficiency, nucleated by the mechanistic target of rapamycin (mTOR), a key regulator of cell growth via modulation of key processes including protein synthesis and autophagy. The precise ways in which altered amino acid management supports cellular transformation remain mostly elusive, and a fuller mechanistic understanding of these processes will be important for efforts to exploit such alterations for cancer therapy.
Keywords: Amino acids; Cancer; Glutamine; Metabolism; Serine; mTOR.
Copyright © 2015. Published by Elsevier Ltd.
Figures



References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

