Label-free quantitative phosphoproteomics with novel pairwise abundance normalization reveals synergistic RAS and CIP2A signaling
- PMID: 26278961
- PMCID: PMC4642524
- DOI: 10.1038/srep13099
Label-free quantitative phosphoproteomics with novel pairwise abundance normalization reveals synergistic RAS and CIP2A signaling
Abstract
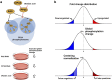
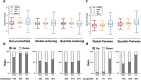
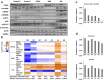
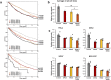
Hyperactivated RAS drives progression of many human malignancies. However, oncogenic activity of RAS is dependent on simultaneous inactivation of protein phosphatase 2A (PP2A) activity. Although PP2A is known to regulate some of the RAS effector pathways, it has not been systematically assessed how these proteins functionally interact. Here we have analyzed phosphoproteomes regulated by either RAS or PP2A, by phosphopeptide enrichment followed by mass-spectrometry-based label-free quantification. To allow data normalization in situations where depletion of RAS or PP2A inhibitor CIP2A causes a large uni-directional change in the phosphopeptide abundance, we developed a novel normalization strategy, named pairwise normalization. This normalization is based on adjusting phosphopeptide abundances measured before and after the enrichment. The superior performance of the pairwise normalization was verified by various independent methods. Additionally, we demonstrate how the selected normalization method influences the downstream analyses and interpretation of pathway activities. Consequently, bioinformatics analysis of RAS and CIP2A regulated phosphoproteomes revealed a significant overlap in their functional pathways. This is most likely biologically meaningful as we observed a synergistic survival effect between CIP2A and RAS expression as well as KRAS activating mutations in TCGA pan-cancer data set, and synergistic relationship between CIP2A and KRAS depletion in colony growth assays.
Figures



References
-
- Zhang J., Yang P. L. & Gray N. S. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 9, 28–39 (2009). - PubMed
-
- Rajalingam K., Schreck R., Rapp U. R. & Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta 1773, 1177–1195 (2007). - PubMed
-
- Haluska F. G. et al.. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res 12, 2301s–2307s (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

