Intramolecular C2 Domain-Mediated Autoinhibition of Protein Kinase C βII
- PMID: 26279568
- PMCID: PMC4551583
- DOI: 10.1016/j.celrep.2015.07.039
Intramolecular C2 Domain-Mediated Autoinhibition of Protein Kinase C βII
Abstract
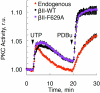
The signaling output of protein kinase C (PKC) is exquisitely controlled, with its disruption resulting in pathophysiologies. Identifying the structural basis for autoinhibition is central to developing effective therapies for cancer, where PKC activity needs to be enhanced, or neurodegenerative diseases, where PKC activity should be inhibited. Here, we reinterpret a previously reported crystal structure of PKCβII and use docking and functional analysis to propose an alternative structure that is consistent with previous literature on PKC regulation. Mutagenesis of predicted contact residues establishes that the Ca(2+)-sensing C2 domain interacts intramolecularly with the kinase domain and the carboxyl-terminal tail, locking PKC in an inactive conformation. Ca(2+)-dependent bridging of the C2 domain to membranes provides the first step in activating PKC via conformational selection. Although the placement of the C1 domains remains to be determined, elucidation of the structural basis for autoinhibition of PKCβII unveils a unique direction for therapeutically targeting PKC.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Banci L, Cavallaro G, Kheifets V, Mochly-Rosen D. Molecular dynamics characterization of the C2 domain of protein kinase Cbeta. The Journal of biological chemistry. 2002;277:12988–12997. - PubMed
-
- Belin RJ, Sumandea MP, Allen EJ, Schoenfelt K, Wang H, Solaro RJ, de Tombe PP. Augmented protein kinase C-alpha-induced myofilament protein phosphorylation contributes to myofilament dysfunction in experimental congestive heart failure. Circulation research. 2007;101:195–204. - PubMed
-
- Bernado P. Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. Eur Biophys J. 2010;39:769–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

