EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy
- PMID: 26280328
- PMCID: PMC4589481
- DOI: 10.1038/nmeth.3541
EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy
Abstract
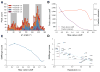
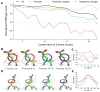
Advances in high-resolution cryo-electron microscopy (cryo-EM) require the development of validation metrics to independently assess map quality and model geometry. We report EMRinger, a tool that assesses the precise fitting of an atomic model into the map during refinement and shows how radiation damage alters scattering from negatively charged amino acids. EMRinger (https://github.com/fraser-lab/EMRinger) will be useful for monitoring progress in resolving and modeling high-resolution features in cryo-EM.
Conflict of interest statement
Figures
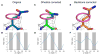
References
-
- Shi Y. Cell. 2014;159:995–1014. - PubMed
-
- Wong W, et al. eLife. 2014;3
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- R21 GM110580/GM/NIGMS NIH HHS/United States
- GM082250/GM/NIGMS NIH HHS/United States
- T32 GM008284/GM/NIGMS NIH HHS/United States
- T32 EB009383/EB/NIBIB NIH HHS/United States
- GM063210/GM/NIGMS NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- P01 GM063210/GM/NIGMS NIH HHS/United States
- OD009180/OD/NIH HHS/United States
- DP5 OD009180/OD/NIH HHS/United States
- GM110580/GM/NIGMS NIH HHS/United States
- R01 GM082893/GM/NIGMS NIH HHS/United States
- T32GM008284/GM/NIGMS NIH HHS/United States
- GM082893/GM/NIGMS NIH HHS/United States
- GM098672/GM/NIGMS NIH HHS/United States
- R01 GM098672/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

