JMS: An Open Source Workflow Management System and Web-Based Cluster Front-End for High Performance Computing
- PMID: 26280450
- PMCID: PMC4539224
- DOI: 10.1371/journal.pone.0134273
JMS: An Open Source Workflow Management System and Web-Based Cluster Front-End for High Performance Computing
Abstract
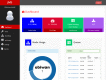
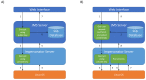
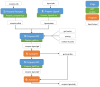
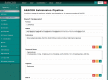
Complex computational pipelines are becoming a staple of modern scientific research. Often these pipelines are resource intensive and require days of computing time. In such cases, it makes sense to run them over high performance computing (HPC) clusters where they can take advantage of the aggregated resources of many powerful computers. In addition to this, researchers often want to integrate their workflows into their own web servers. In these cases, software is needed to manage the submission of jobs from the web interface to the cluster and then return the results once the job has finished executing. We have developed the Job Management System (JMS), a workflow management system and web interface for high performance computing (HPC). JMS provides users with a user-friendly web interface for creating complex workflows with multiple stages. It integrates this workflow functionality with the resource manager, a tool that is used to control and manage batch jobs on HPC clusters. As such, JMS combines workflow management functionality with cluster administration functionality. In addition, JMS provides developer tools including a code editor and the ability to version tools and scripts. JMS can be used by researchers from any field to build and run complex computational pipelines and provides functionality to include these pipelines in external interfaces. JMS is currently being used to house a number of bioinformatics pipelines at the Research Unit in Bioinformatics (RUBi) at Rhodes University. JMS is an open-source project and is freely available at https://github.com/RUBi-ZA/JMS.
Conflict of interest statement
Figures
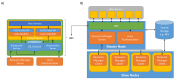

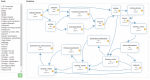
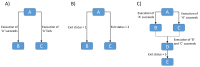


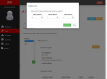



References
-
- Adaptive Computing Enterprises Inc. Torque [Internet]. 2015. Available: http://www.adaptivecomputing.com/products/open-source/torque/
-
- Jette M, Grondona M. SLURM: Simple Linux Utility for Resource Management. ClusterWorld Conference and Expo CWCE. 2003. pp. 44–60. 10.1007/10968987 - DOI
-
- Jackson D, Snell Q, Clement M. Core Algorithms of the Maui Scheduler. Job Sched Strateg Parallel Process. 2001;2221: 87–102. 10.1007/3-540-45540-X_6 - DOI
-
- Misra G, Agrawal S, Kurkure N, Pawar S, Mathur K. CHReME: A Web Based Application Execution Tool for using HPC Resources. International Conference on High Performance Computing. 2011. pp. 12–14.
-
- Adaptive Computing Enterprises Inc. Adaptive Computing products [Internet]. 2015. Available: http://www.adaptivecomputing.com/products/hpc-products/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

