Variability and pathogenicity of hepatitis E virus genotype 3 variants
- PMID: 26282123
- PMCID: PMC4806580
- DOI: 10.1099/jgv.0.000264
Variability and pathogenicity of hepatitis E virus genotype 3 variants
Abstract
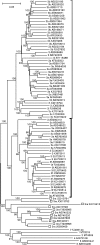
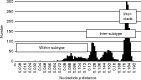
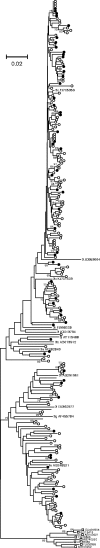
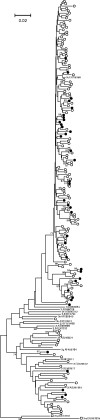
Infection with hepatitis E virus (HEV) can be clinically inapparent or produce symptoms and signs of hepatitis of varying severity and occasional fatality. This variability in clinical outcomes may reflect differences in host susceptibility or the presence of virally encoded determinants of pathogenicity. Analysis of complete genome sequences supports the division of HEV genotype 3 (HEV-3) variants into three major clades: 3ra comprising HEV isolates from rabbits, and 3efg and 3abchij comprising the corresponding named subtypes derived from humans and pigs. Using this framework, we investigated associations between viral genetic variability of HEV-3 in symptomatic and asymptomatic infections by comparing HEV-3 subgenomic sequences previously obtained from blood donors with those from patients presenting with hepatitis in the UK (54 blood donors, 148 hepatitis patients), the Netherlands (38 blood donors, 119 hepatitis patients), France (24 blood donors, 55 hepatitis patients) and Germany (14 blood donors, 36 hepatitis patients). In none of these countries was evidence found for a significant association between virus variants and patient group (P>0.05 Fisher's exact test). Furthermore, within a group of 123 patients in Scotland with clinically apparent HEV infections, we found no evidence for an association between variants of HEV-3 and disease severity or alanine aminotransferase level. The lack of detectable virally encoded determinants of disease outcomes in HEV-3 infection implies a more important role for host factors in its clinical phenotype.
Figures

References
-
- Abe T., Aikawa T., Akahane Y., Arai M., Asahina Y., Atarashi Y., Chayama K., Harada H., Hashimoto N., other authors (2006). [Demographic, epidemiological, and virological characteristics of hepatitis E virus infections in Japan based on 254 human cases collected nationwide] Kanzo 47 384–391 (in Japanese with English abstract).
-
- Bu Q., Wang X., Wang L., Liu P., Geng J., Wang M., Han J., Zhu Y., Zhuang H. (2013). Hepatitis E virus genotype 4 isolated from a patient with liver failure: full-length sequence analysis showing potential determinants of virus pathogenesis Arch Virol 158 165–172 10.1007/s00705-012-1488-3 . - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

