Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains
- PMID: 26282408
- PMCID: PMC4604379
- DOI: 10.1128/AAC.00952-15
Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains
Abstract
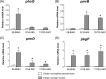
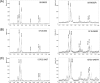
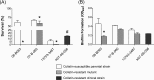
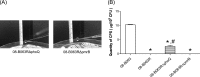
In this study, we investigated the effects of colistin resistance on virulence and fitness in hypermucoviscous (HV) Klebsiella pneumoniae sequence type 23 (ST23) strains. Colistin-resistant mutants were developed from three colistin-susceptible HV K. pneumoniae ST23 strains. The lipid A structures of strains were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Changes in HV were investigated using the string test, and extracellular polysaccharide production was quantified. The expression levels of the phoQ, pmrD, pmrB, pbgP, magA, and p-rmpA2 genes, serum resistance, and biofilm-forming activity were determined. The fitness of colistin-resistant mutants compared to that of the parental strains was examined by determining the competitive index (CI). The colistin-resistant mutants exhibited reduced HV, which was accompanied by decreased formation of capsular polysaccharides (CPS) and reduced expression of genes (magA and p-rmpA2). While there was enhanced expression of pmrD and pbgP in all colistin-resistant derivatives, there were differences in the expression levels of phoQ and pmrB between strains. MALDI-TOF analysis detected the addition of aminoarabinose or palmitate to the lipid A moiety of lipopolysaccharide in the colistin-resistant derivatives. In addition, survival rates in the presence of normal human serum were decreased in the mutant strains, and CI values (0.01 to 0.19) indicated significant fitness defects in the colistin-resistant derivatives compared to the respective parental strains. In hypervirulent HV K. pneumoniae strains, the acquisition of colistin resistance was accompanied by reduced CPS production, impaired virulence, and a significant fitness cost.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, Ip M. 2011. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol 49:3761–3765. doi:10.1128/JCM.00977-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

