Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis
- PMID: 26283368
- PMCID: PMC4568209
- DOI: 10.1073/pnas.1507760112
Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis
Abstract
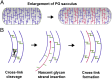
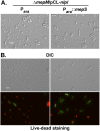
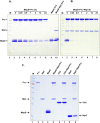
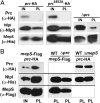
Bacterial growth and morphogenesis are intimately coupled to expansion of peptidoglycan (PG), an extensively cross-linked macromolecule that forms a protective mesh-like sacculus around the cytoplasmic membrane. Growth of the PG sacculus is a dynamic event requiring the concerted action of hydrolases that cleave the cross-links for insertion of new material and synthases that catalyze cross-link formation; however, the factors that regulate PG expansion during bacterial growth are poorly understood. Here, we show that the PG hydrolase MepS (formerly Spr), which is specific to cleavage of cross-links during PG expansion in Escherichia coli, is modulated by proteolysis. Using combined genetic, molecular, and biochemical approaches, we demonstrate that MepS is rapidly degraded by a proteolytic system comprising an outer membrane lipoprotein of unknown function, NlpI, and a periplasmic protease, Prc (or Tsp). In summary, our results indicate that the NlpI-Prc system contributes to growth and enlargement of the PG sacculus by modulating the cellular levels of the cross-link-cleaving hydrolase MepS. Overall, this study signifies the importance of PG cross-link cleavage and its regulation in bacterial cell wall biogenesis.
Keywords: MepS; NlpI-Prc; bacterial morphogenesis; peptidoglycan; regulated proteolysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








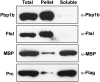

References
-
- Weidel W, Pelzer H. Bagshaped macromolecules: A new outlook on bacterial cell walls. Adv Enzymol Relat Areas Mol Biol. 1964;26:193–232. - PubMed
-
- Park JT. The murein sacculus. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. ASM; Washington, DC: 1996. pp. 48–57.
-
- den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32(2):321–344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

