Mechanism, factors, and physiological role of nonsense-mediated mRNA decay
- PMID: 26283621
- PMCID: PMC11113733
- DOI: 10.1007/s00018-015-2017-9
Mechanism, factors, and physiological role of nonsense-mediated mRNA decay
Abstract
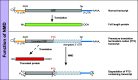
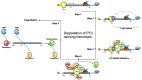
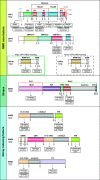
Nonsense-mediated mRNA decay (NMD) is a translation-dependent, multistep process that degrades irregular or faulty messenger RNAs (mRNAs). NMD mainly targets mRNAs with a truncated open reading frame (ORF) due to premature termination codons (PTCs). In addition, NMD also regulates the expression of different types of endogenous mRNA substrates. A multitude of factors are involved in the tight regulation of the NMD mechanism. In this review, we focus on the molecular mechanism of mammalian NMD. Based on the published data, we discuss the involvement of translation termination in NMD initiation. Furthermore, we provide a detailed overview of the core NMD machinery, as well as several peripheral NMD factors, and discuss their function. Finally, we present an overview of diseases associated with NMD factor mutations and summarize the current state of treatment for genetic disorders caused by nonsense mutations.
Keywords: Exon junction complex; Genetic disease; NMD; Quality control; UPF1.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

