TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells
- PMID: 26283799
- PMCID: PMC4539992
- DOI: 10.1083/jcb.201502107
TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells
Abstract
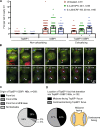
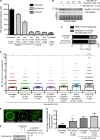
Genome integrity is critically dependent on timely DNA replication and accurate chromosome segregation. Replication stress delays replication into G2/M, which in turn impairs proper chromosome segregation and inflicts DNA damage on the daughter cells. Here we show that TopBP1 forms foci upon mitotic entry. In early mitosis, TopBP1 marks sites of and promotes unscheduled DNA synthesis. Moreover, TopBP1 is required for focus formation of the structure-selective nuclease and scaffold protein SLX4 in mitosis. Persistent TopBP1 foci transition into 53BP1 nuclear bodies (NBs) in G1 and precise temporal depletion of TopBP1 just before mitotic entry induced formation of 53BP1 NBs in the next cell cycle, showing that TopBP1 acts to reduce transmission of DNA damage to G1 daughter cells. Based on these results, we propose that TopBP1 maintains genome integrity in mitosis by controlling chromatin recruitment of SLX4 and by facilitating unscheduled DNA synthesis.
© 2015 Pedersen et al.
Figures







References
-
- Bergoglio V., Boyer A.S., Walsh E., Naim V., Legube G., Lee M.Y., Rey L., Rosselli F., Cazaux C., Eckert K.A., and Hoffmann J.S.. 2013. DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J. Cell Biol. 201:395–408. 10.1083/jcb.201207066 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

