NADPH-generating systems in bacteria and archaea
- PMID: 26284036
- PMCID: PMC4518329
- DOI: 10.3389/fmicb.2015.00742
NADPH-generating systems in bacteria and archaea
Abstract
Reduced nicotinamide adenine dinucleotide phosphate (NADPH) is an essential electron donor in all organisms. It provides the reducing power that drives numerous anabolic reactions, including those responsible for the biosynthesis of all major cell components and many products in biotechnology. The efficient synthesis of many of these products, however, is limited by the rate of NADPH regeneration. Hence, a thorough understanding of the reactions involved in the generation of NADPH is required to increase its turnover through rational strain improvement. Traditionally, the main engineering targets for increasing NADPH availability have included the dehydrogenase reactions of the oxidative pentose phosphate pathway and the isocitrate dehydrogenase step of the tricarboxylic acid (TCA) cycle. However, the importance of alternative NADPH-generating reactions has recently become evident. In the current review, the major canonical and non-canonical reactions involved in the production and regeneration of NADPH in prokaryotes are described, and their key enzymes are discussed. In addition, an overview of how different enzymes have been applied to increase NADPH availability and thereby enhance productivity is provided.
Keywords: GAPN; NADPH regeneration; ferredoxin:NADP+ oxidoreductase; hydrogenase; isocitrate dehydrogenase; malic enzyme; pentose phosphate pathway; transhydrogenase.
Figures
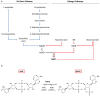
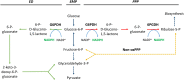
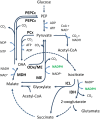
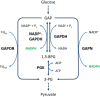

References
-
- Adachi O., Ameyama M. (1981). d-Glucose dehydrogenase from Gluconobacter suboxydans: solubilization, purification and characterization. Agric. Biol. Chem. 45, 159–163.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

