Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows
- PMID: 26284058
- PMCID: PMC4521595
- DOI: 10.3389/fmicb.2015.00781
Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows
Abstract
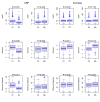
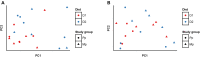
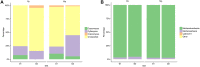
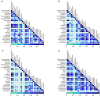
The rumen microbiome represents a complex microbial genetic web where bacteria, anaerobic rumen fungi (ARF), protozoa and archaea work in harmony contributing to the health and productivity of ruminants. We hypothesized that the rumen microbiome shifts as the dairy cow advances in lactations and these microbial changes may contribute to differences in productivity between primiparous (first lactation) and multiparous (≥second lactation) cows. To this end, we investigated shifts in the ruminal ARF and methanogenic communities in both primiparous (n = 5) and multiparous (n = 5) cows as they transitioned from a high forage to a high grain diet upon initiation of lactation. A total of 20 rumen samples were extracted for genomic DNA, amplified using archaeal and fungal specific primers, sequenced on a 454 platform and analyzed using QIIME. Community comparisons (Bray-Curtis index) revealed the effect of diet (P < 0.01) on ARF composition, while archaeal communities differed between primiparous and multiparous cows (P < 0.05). Among ARF, several lineages were unclassified, however, phylum Neocallimastigomycota showed the presence of three known genera. Abundance of Cyllamyces and Caecomyces shifted with diet, whereas Orpinomyces was influenced by both diet and age. Methanobrevibacter constituted the most dominant archaeal genus across all samples. Co-occurrence analysis incorporating taxa from bacteria, ARF and archaea revealed syntrophic interactions both within and between microbial domains in response to change in diet as well as age of dairy cows. Notably, these interactions were numerous and complex in multiparous cows, supporting our hypothesis that the rumen microbiome also matures with age to sustain the growing metabolic needs of the host. This study provides a broader picture of the ARF and methanogenic populations in the rumen of dairy cows and their co-occurrence implicates specific relationships between different microbial domains in response to diet and age.
Keywords: anaerobic fungi; co-occurrence; diet; methanogenic archaea; microbiome; rumen.
Figures
References
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26 32–46.
LinkOut - more resources
Full Text Sources
Other Literature Sources

