Microbial Communities Can Be Described by Metabolic Structure: A General Framework and Application to a Seasonally Variable, Depth-Stratified Microbial Community from the Coastal West Antarctic Peninsula
- PMID: 26285202
- PMCID: PMC4540456
- DOI: 10.1371/journal.pone.0135868
Microbial Communities Can Be Described by Metabolic Structure: A General Framework and Application to a Seasonally Variable, Depth-Stratified Microbial Community from the Coastal West Antarctic Peninsula
Abstract
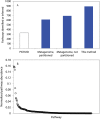
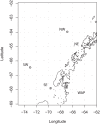
Taxonomic marker gene studies, such as the 16S rRNA gene, have been used to successfully explore microbial diversity in a variety of marine, terrestrial, and host environments. For some of these environments long term sampling programs are beginning to build a historical record of microbial community structure. Although these 16S rRNA gene datasets do not intrinsically provide information on microbial metabolism or ecosystem function, this information can be developed by identifying metabolisms associated with related, phenotyped strains. Here we introduce the concept of metabolic inference; the systematic prediction of metabolism from phylogeny, and describe a complete pipeline for predicting the metabolic pathways likely to be found in a collection of 16S rRNA gene phylotypes. This framework includes a mechanism for assigning confidence to each metabolic inference that is based on a novel method for evaluating genomic plasticity. We applied this framework to 16S rRNA gene libraries from the West Antarctic Peninsula marine environment, including surface and deep summer samples and surface winter samples. Using statistical methods commonly applied to community ecology data we found that metabolic structure differed between summer surface and winter and deep samples, comparable to an analysis of community structure by 16S rRNA gene phylotypes. While taxonomic variance between samples was primarily driven by low abundance taxa, metabolic variance was attributable to both high and low abundance pathways. This suggests that clades with a high degree of functional redundancy can occupy distinct adjacent niches. Overall our findings demonstrate that inferred metabolism can be used in place of taxonomy to describe the structure of microbial communities. Coupling metabolic inference with targeted metagenomics and an improved collection of completed genomes could be a powerful way to analyze microbial communities in a high-throughput manner that provides direct access to metabolic and ecosystem function.
Conflict of interest statement
Figures





References
-
- Martens K, Segers H. Taxonomy and systematics in biodiversity research. Hydrobiologia. 2005;542: 27–31. 10.1007/s10750-005-0892-z - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

