Assessment of Artificial MiRNA Architectures for Higher Knockdown Efficiencies without the Undesired Effects in Mice
- PMID: 26285215
- PMCID: PMC4540464
- DOI: 10.1371/journal.pone.0135919
Assessment of Artificial MiRNA Architectures for Higher Knockdown Efficiencies without the Undesired Effects in Mice
Abstract
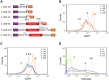
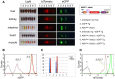
RNAi-based strategies have been used for hypomorphic analyses. However, there are technical challenges to achieve robust, reproducible knockdown effect. Here we examined the artificial microRNA (amiRNA) architectures that could provide higher knockdown efficiencies. Using transient and stable transfection assays in cells, we found that simple amiRNA-expression cassettes, that did not contain a marker gene (-MG), displayed higher amiRNA expression and more efficient knockdown than those that contained a marker gene (+MG). Further, we tested this phenomenon in vivo, by analyzing amiRNA-expressing mice that were produced by the pronuclear injection-based targeted transgenesis (PITT) method. While we observed significant silencing of the target gene (eGFP) in +MG hemizygous mice, obtaining -MG amiRNA expression mice, even hemizygotes, was difficult and the animals died perinatally. We obtained only mosaic mice having both "-MG amiRNA" cells and "amiRNA low-expression" cells but they exhibited growth retardation and cataracts, and they could not transmit the -MG amiRNA allele to the next generation. Furthermore, +MG amiRNA homozygotes could not be obtained. These results suggested that excessive amiRNAs transcribed by -MG expression cassettes cause deleterious effects in mice, and the amiRNA expression level in hemizygous +MG amiRNA mice is near the upper limit, where mice can develop normally. In conclusion, the PITT-(+MG amiRNA) system demonstrated here can generate knockdown mouse models that reliably express highest and tolerable levels of amiRNAs.
Conflict of interest statement
Figures


Similar articles
-
Inhibition of porcine reproductive and respiratory syndrome virus infection by recombinant adenovirus- and/or exosome-delivered the artificial microRNAs targeting sialoadhesin and CD163 receptors.Virol J. 2014 Dec 19;11:225. doi: 10.1186/s12985-014-0225-9. Virol J. 2014. PMID: 25522782 Free PMC article.
-
CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA.Sci Rep. 2015 Aug 5;5:12799. doi: 10.1038/srep12799. Sci Rep. 2015. PMID: 26242611 Free PMC article.
-
Goat activin receptor type IIB knockdown by artificial microRNAs in vitro.Appl Biochem Biotechnol. 2014 Sep;174(1):424-36. doi: 10.1007/s12010-014-1071-3. Epub 2014 Jul 31. Appl Biochem Biotechnol. 2014. PMID: 25080379
-
Scaffolds for Artificial miRNA Expression in Animal Cells.Hum Gene Ther Methods. 2015 Oct;26(5):162-74. doi: 10.1089/hgtb.2015.043. Epub 2015 Sep 25. Hum Gene Ther Methods. 2015. PMID: 26406928 Review.
-
Artificial microRNAs (amiRNAs) engineering - On how microRNA-based silencing methods have affected current plant silencing research.Biochem Biophys Res Commun. 2011 Mar 18;406(3):315-9. doi: 10.1016/j.bbrc.2011.02.045. Epub 2011 Feb 15. Biochem Biophys Res Commun. 2011. PMID: 21329663 Review.
Cited by
-
Targeted insertion of conditional expression cassettes into the mouse genome using the modified i-PITT.BMC Genomics. 2024 Jun 5;25(1):568. doi: 10.1186/s12864-024-10250-0. BMC Genomics. 2024. PMID: 38840068 Free PMC article.
-
Conditional knockout of Foxc2 gene in kidney: efficient generation of conditional alleles of single-exon gene by double-selection system.Mamm Genome. 2016 Feb;27(1-2):62-9. doi: 10.1007/s00335-015-9610-y. Epub 2015 Nov 5. Mamm Genome. 2016. PMID: 26542959
-
Oocyte-specific gene knockdown by intronic artificial microRNAs driven by Zp3 transcription in mice.J Reprod Dev. 2021 Jun 21;67(3):229-234. doi: 10.1262/jrd.2020-146. Epub 2021 Mar 13. J Reprod Dev. 2021. PMID: 33716236 Free PMC article.
-
Optimizing Synthetic miRNA Minigene Architecture for Efficient miRNA Hairpin Concatenation and Multi-target Gene Knockdown.Mol Ther Nucleic Acids. 2019 Mar 1;14:351-363. doi: 10.1016/j.omtn.2018.12.004. Epub 2018 Dec 14. Mol Ther Nucleic Acids. 2019. PMID: 30665184 Free PMC article.
-
Pronuclear Injection-Based Targeted Transgenesis.Curr Protoc Hum Genet. 2016 Oct 11;91:15.10.1-15.10.28. doi: 10.1002/cphg.23. Curr Protoc Hum Genet. 2016. PMID: 27727435 Free PMC article.
References
-
- Mittal V. Improving the efficiency of RNA interference in mammals. Nat Rev Genet. 2004;5(5):355–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

