4-protein signature predicting tamoxifen treatment outcome in recurrent breast cancer
- PMID: 26285647
- PMCID: PMC5528925
- DOI: 10.1016/j.molonc.2015.07.004
4-protein signature predicting tamoxifen treatment outcome in recurrent breast cancer
Abstract
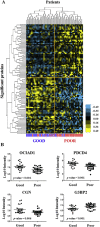
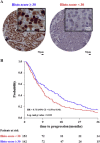
Estrogen receptor (ER) positive tumors represent the majority of breast malignancies, and are effectively treated with hormonal therapies, such as tamoxifen. However, in the recurrent disease resistance to tamoxifen therapy is common and a major cause of death. In recent years, in-depth proteome analyses have enabled identification of clinically useful biomarkers, particularly, when heterogeneity in complex tumor tissue was reduced using laser capture microdissection (LCM). In the current study, we performed high resolution proteomic analysis on two cohorts of ER positive breast tumors derived from patients who either manifested good or poor outcome to tamoxifen treatment upon recurrence. A total of 112 fresh frozen tumors were collected from multiple medical centers and divided into two sets: an in-house training and a multi-center test set. Epithelial tumor cells were enriched with LCM and analyzed by nano-LC Orbitrap mass spectrometry (MS), which yielded >3000 and >4000 quantified proteins in the training and test sets, respectively. Raw data are available via ProteomeXchange with identifiers PXD000484 and PXD000485. Statistical analysis showed differential abundance of 99 proteins, of which a subset of 4 proteins was selected through a multivariate step-down to develop a predictor for tamoxifen treatment outcome. The 4-protein signature significantly predicted poor outcome patients in the test set, independent of predictive histopathological characteristics (hazard ratio [HR] = 2.17; 95% confidence interval [CI] = 1.15 to 4.17; multivariate Cox regression p value = 0.017). Immunohistochemical (IHC) staining of PDCD4, one of the signature proteins, on an independent set of formalin-fixed paraffin-embedded tumor tissues provided and independent technical validation (HR = 0.72; 95% CI = 0.57 to 0.92; multivariate Cox regression p value = 0.009). We hereby report the first validated protein predictor for tamoxifen treatment outcome in recurrent ER-positive breast cancer. IHC further showed that PDCD4 is an independent marker.
Keywords: Biomarker; Breast cancer; Mass spectrometry; Proteomics; Tamoxifen resistance.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

