Identification of inhibitors that target dual-specificity phosphatase 5 provide new insights into the binding requirements for the two phosphate pockets
- PMID: 26286528
- PMCID: PMC4545774
- DOI: 10.1186/s12858-015-0048-3
Identification of inhibitors that target dual-specificity phosphatase 5 provide new insights into the binding requirements for the two phosphate pockets
Abstract
Background: Dual-specificity phosphatase-5 (DUSP5) plays a central role in vascular development and disease. We present a p-nitrophenol phosphate (pNPP) based enzymatic assay to screen for inhibitors of the phosphatase domain of DUSP5.
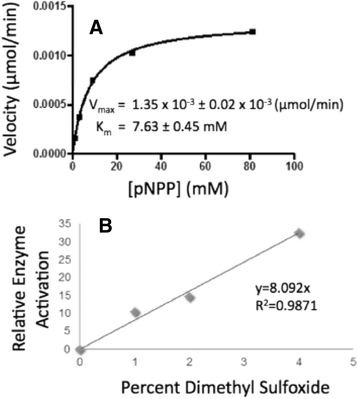
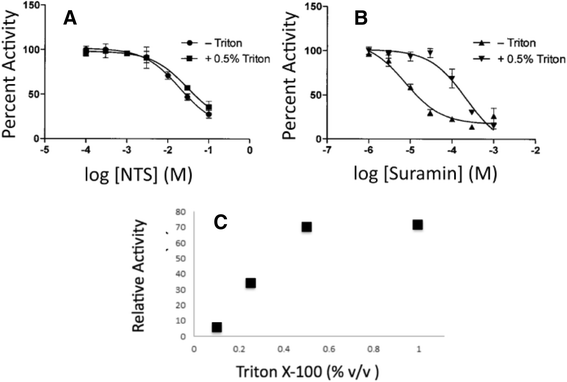
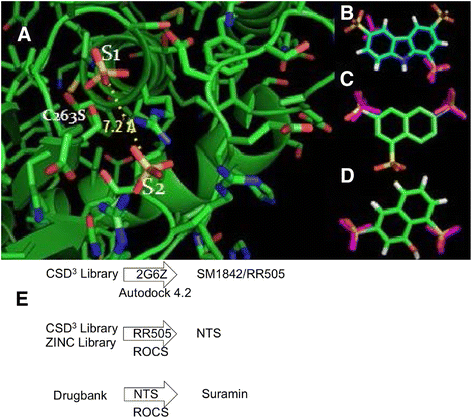
Methods: pNPP is a mimic of the phosphorylated tyrosine on the ERK2 substrate (pERK2) and binds the DUSP5 phosphatase domain with a Km of 7.6 ± 0.4 mM. Docking followed by inhibitor verification using the pNPP assay identified a series of polysulfonated aromatic inhibitors that occupy the DUSP5 active site in the region that is likely occupied by the dual-phosphorylated ERK2 substrate tripeptide (pThr-Glu-pTyr). Secondary assays were performed with full length DUSP5 with ERK2 as substrate.
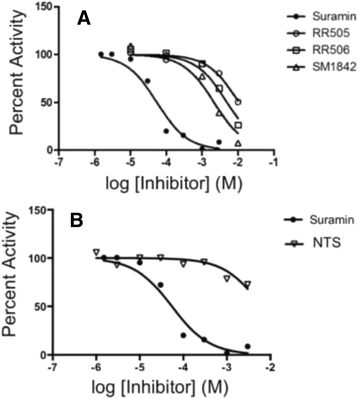
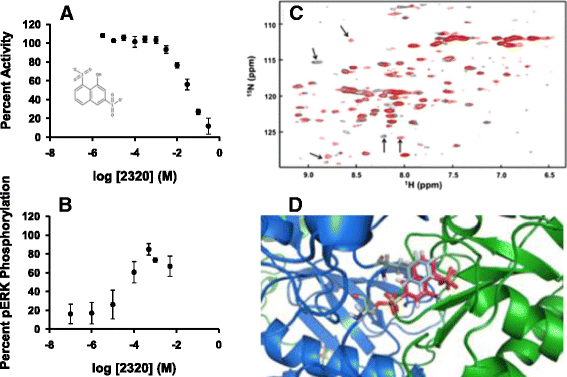
Results: The most potent inhibitor has a naphthalene trisulfonate (NTS) core. A search for similar compounds in a drug database identified suramin, a dimerized form of NTS. While suramin appears to be a potent and competitive inhibitor (25 ± 5 μM), binding to the DUSP5 phosphatase domain more tightly than the monomeric ligands of which it is comprised, it also aggregates. Further ligand-based screening, based on a pharmacophore derived from the 7 Å separation of sulfonates on inhibitors and on sulfates present in the DUSP5 crystal structure, identified a disulfonated and phenolic naphthalene inhibitor (CSD (3) _2320) with IC₅₀ of 33 μM that is similar to NTS and does not aggregate.
Conclusions: The new DUSP5 inhibitors we identify in this study typically have sulfonates 7 Å apart, likely positioning them where the two phosphates of the substrate peptide (pThr-Glu-pTyr) bind, with one inhibitor also positioning a phenolic hydroxyl where the water nucleophile may reside. Polysulfonated aromatic compounds do not commonly appear in drugs and have a tendency to aggregate. One FDA-approved polysulfonated drug, suramin, inhibits DUSP5 and also aggregates. Docking and modeling studies presented herein identify polysulfonated aromatic inhibitors that do not aggregate, and provide insights to guide future design of mimics of the dual-phosphate loops of the ERK substrates for DUSPs.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Research Materials
Miscellaneous

