Read clouds uncover variation in complex regions of the human genome
- PMID: 26286554
- PMCID: PMC4579342
- DOI: 10.1101/gr.191189.115
Read clouds uncover variation in complex regions of the human genome
Abstract
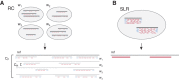
Although an increasing amount of human genetic variation is being identified and recorded, determining variants within repeated sequences of the human genome remains a challenge. Most population and genome-wide association studies have therefore been unable to consider variation in these regions. Core to the problem is the lack of a sequencing technology that produces reads with sufficient length and accuracy to enable unique mapping. Here, we present a novel methodology of using read clouds, obtained by accurate short-read sequencing of DNA derived from long fragment libraries, to confidently align short reads within repeat regions and enable accurate variant discovery. Our novel algorithm, Random Field Aligner (RFA), captures the relationships among the short reads governed by the long read process via a Markov Random Field. We utilized a modified version of the Illumina TruSeq synthetic long-read protocol, which yielded shallow-sequenced read clouds. We test RFA through extensive simulations and apply it to discover variants on the NA12878 human sample, for which shallow TruSeq read cloud sequencing data are available, and on an invasive breast carcinoma genome that we sequenced using the same method. We demonstrate that RFA facilitates accurate recovery of variation in 155 Mb of the human genome, including 94% of 67 Mb of segmental duplication sequence and 96% of 11 Mb of transcribed sequence, that are currently hidden from short-read technologies.
© 2015 Bishara et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O'Grady J. 2014. Minion nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 33: 296–300. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
