Performance evaluation of indel calling tools using real short-read data
- PMID: 26286629
- PMCID: PMC4545535
- DOI: 10.1186/s40246-015-0042-2
Performance evaluation of indel calling tools using real short-read data
Abstract
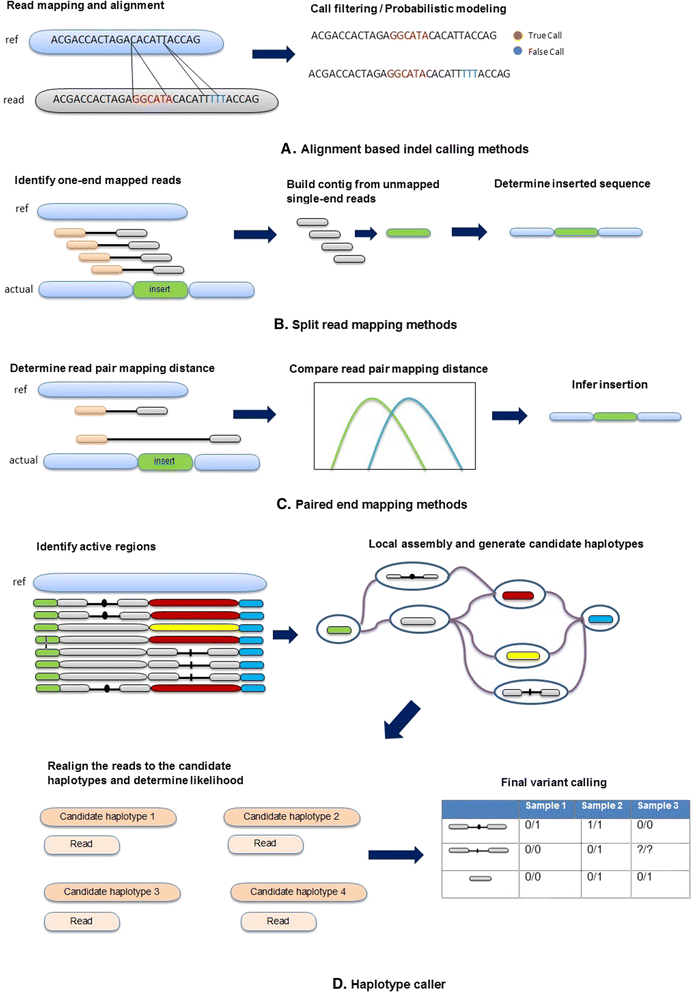
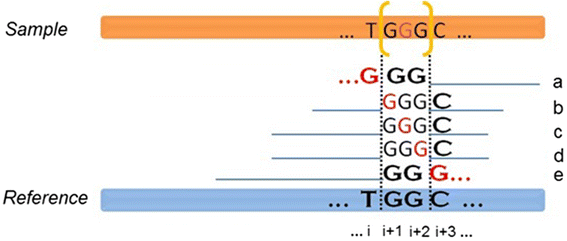
Background: Insertion and deletion (indel), a common form of genetic variation, has been shown to cause or contribute to human genetic diseases and cancer. With the advance of next-generation sequencing technology, many indel calling tools have been developed; however, evaluation and comparison of these tools using large-scale real data are still scant. Here we evaluated seven popular and publicly available indel calling tools, GATK Unified Genotyper, VarScan, Pindel, SAMtools, Dindel, GTAK HaplotypeCaller, and Platypus, using 78 human genome low-coverage data from the 1000 Genomes project.
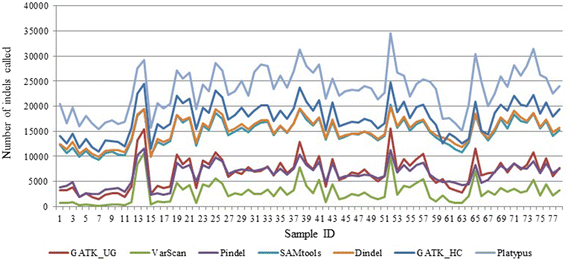
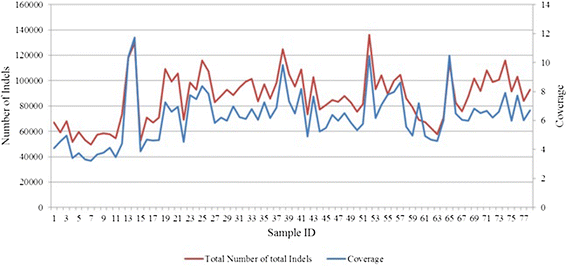
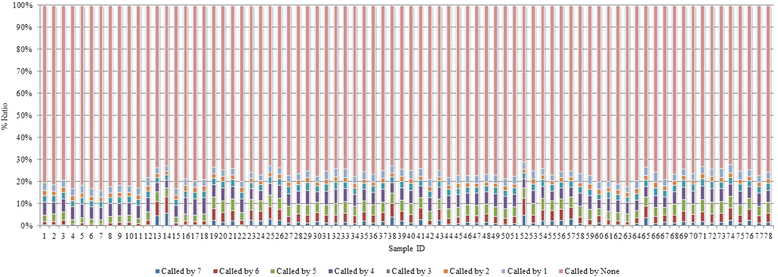
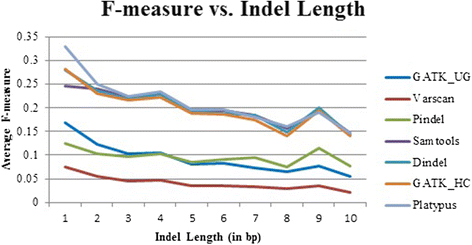
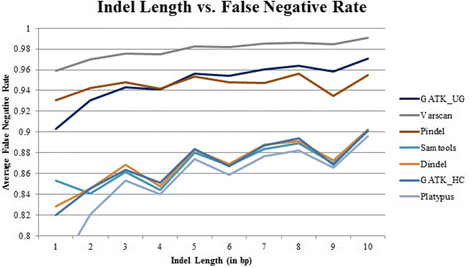
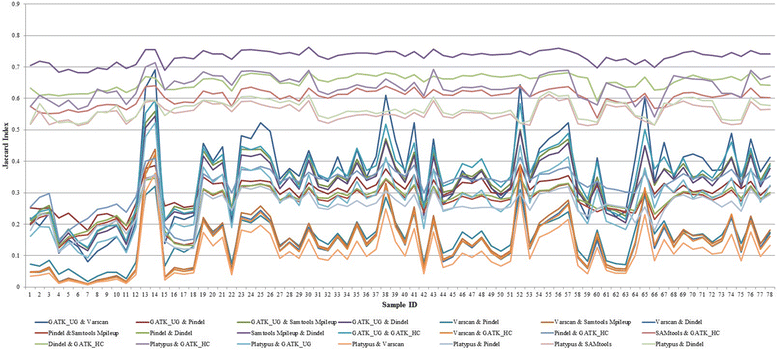
Results: Comparing indels called by these tools with a known set of indels, we found that Platypus outperforms other tools. In addition, a high percentage of known indels still remain undetected and the number of common indels called by all seven tools is very low.
Conclusion: All these findings indicate the necessity of improving the existing tools or developing new algorithms to achieve reliable and consistent indel calling results.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

