Convergent Evolution of Endosymbiont Differentiation in Dalbergioid and Inverted Repeat-Lacking Clade Legumes Mediated by Nodule-Specific Cysteine-Rich Peptides
- PMID: 26286718
- PMCID: PMC4587450
- DOI: 10.1104/pp.15.00584
Convergent Evolution of Endosymbiont Differentiation in Dalbergioid and Inverted Repeat-Lacking Clade Legumes Mediated by Nodule-Specific Cysteine-Rich Peptides
Abstract
Nutritional symbiotic interactions require the housing of large numbers of microbial symbionts, which produce essential compounds for the growth of the host. In the legume-rhizobium nitrogen-fixing symbiosis, thousands of rhizobium microsymbionts, called bacteroids, are confined intracellularly within highly specialized symbiotic host cells. In Inverted Repeat-Lacking Clade (IRLC) legumes such as Medicago spp., the bacteroids are kept under control by an arsenal of nodule-specific cysteine-rich (NCR) peptides, which induce the bacteria in an irreversible, strongly elongated, and polyploid state. Here, we show that in Aeschynomene spp. legumes belonging to the more ancient Dalbergioid lineage, bacteroids are elongated or spherical depending on the Aeschynomene spp. and that these bacteroids are terminally differentiated and polyploid, similar to bacteroids in IRLC legumes. Transcriptome, in situ hybridization, and proteome analyses demonstrated that the symbiotic cells in the Aeschynomene spp. nodules produce a large diversity of NCR-like peptides, which are transported to the bacteroids. Blocking NCR transport by RNA interference-mediated inactivation of the secretory pathway inhibits bacteroid differentiation. Together, our results support the view that bacteroid differentiation in the Dalbergioid clade, which likely evolved independently from the bacteroid differentiation in the IRLC clade, is based on very similar mechanisms used by IRLC legumes.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures
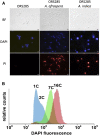
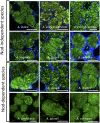

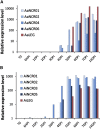

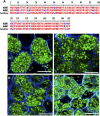
References
-
- Alunni B, Kevei Z, Redondo-Nieto M, Kondorosi A, Mergaert P, Kondorosi E (2007) Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol Plant Microbe Interact 20: 1138–1148 - PubMed
-
- Arrighi JF, Cartieaux F, Brown SC, Rodier-Goud M, Boursot M, Fardoux J, Patrel D, Gully D, Fabre S, Chaintreuil C, et al. (2012) Aeschynomene evenia, a model plant for studying the molecular genetics of the nod-independent rhizobium-legume symbiosis. Mol Plant Microbe Interact 25: 851–861 - PubMed
-
- Arrighi JF, Chaintreuil C, Cartieaux F, Cardi C, Rodier-Goud M, Brown SC, Boursot M, D’Hont A, Dreyfus B, Giraud E (2014) Radiation of the Nod-independent Aeschynomene relies on multiple allopolyploid speciation events. New Phytol 201: 1457–1468 - PubMed
-
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al. (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 - PubMed
-
- Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N, Goormachtig S, Giraud E (2011) Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol Plant Microbe Interact 24: 1359–1371 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

