Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database
- PMID: 26286858
- PMCID: PMC4602074
- DOI: 10.1007/s00335-015-9595-6
Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database
Abstract
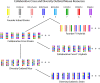
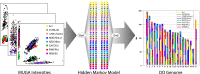
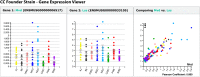
The Mouse Phenome Database was originally conceived as a platform for the integration of phenotype data collected on a defined collection of 40 inbred mouse strains--the "phenome panel." This model provided an impetus for community data sharing, and integration was readily achieved through the reproducible genotypes of the phenome panel strains. Advances in the development of mouse populations lead to an expanded role of the Mouse Phenome Database to encompass new strain panels and inbred strain crosses. The recent introduction of the Collaborative Cross and Diversity Outbred mice, which share an extensive pool of genetic variation from eight founder inbred strains, presents new opportunities and challenges for community data resources. A wide variety of molecular and clinical phenotypes are being collected across genotypes, tissues, ages, environmental exposures, interventions, and treatments. The Mouse Phenome Database provides a framework for retrieval, integration, analysis, and display of these data, enabling them to be evaluated in the context of existing data from standard inbred strains. Primary data in the Mouse Phenome Database are supported by extensive metadata on protocols and procedures. These are centrally curated to ensure accuracy and reproducibility and to provide data in consistent formats. The Mouse Phenome Database represents an established and growing community data resource for mouse phenotype data and encourages submissions from new mouse resources, enabling investigators to integrate existing data into their studies of the phenotypic consequences of genetic variation.
Figures



References
-
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. - DOI - PMC - PubMed
-
- Bubier JA, Jay JJ, Baker CL, Bergeson SE, Ohno H, Metten P, Crabbe JC, Chesler EJ. Identification of a QTL in Mus musculus for alcohol preference, withdrawal, and Ap3m2 expression using integrative functional genomics and precision genetics. Genetics. 2014;197:1377–1393. doi: 10.1534/genetics.114.166165. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

