Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression
- PMID: 26286985
- PMCID: PMC4547249
- DOI: 10.1073/pnas.1500012112
Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression
Abstract
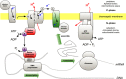
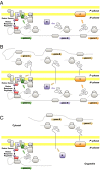
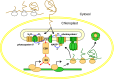
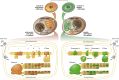
Chloroplasts and mitochondria are subcellular bioenergetic organelles with their own genomes and genetic systems. DNA replication and transmission to daughter organelles produces cytoplasmic inheritance of characters associated with primary events in photosynthesis and respiration. The prokaryotic ancestors of chloroplasts and mitochondria were endosymbionts whose genes became copied to the genomes of their cellular hosts. These copies gave rise to nuclear chromosomal genes that encode cytosolic proteins and precursor proteins that are synthesized in the cytosol for import into the organelle into which the endosymbiont evolved. What accounts for the retention of genes for the complete synthesis within chloroplasts and mitochondria of a tiny minority of their protein subunits? One hypothesis is that expression of genes for protein subunits of energy-transducing enzymes must respond to physical environmental change by means of a direct and unconditional regulatory control--control exerted by change in the redox state of the corresponding gene product. This hypothesis proposes that, to preserve function, an entire redox regulatory system has to be retained within its original membrane-bound compartment. Colocation of gene and gene product for redox regulation of gene expression (CoRR) is a hypothesis in agreement with the results of a variety of experiments designed to test it and which seem to have no other satisfactory explanation. Here, I review evidence relating to CoRR and discuss its development, conclusions, and implications. This overview also identifies predictions concerning the results of experiments that may yet prove the hypothesis to be incorrect.
Keywords: CoRR hypothesis; chloroplast; mitochondrion; oxidative phosphorylation; photosynthesis.
Conflict of interest statement
The author declares no conflict of interest.
Figures
References
-
- Allen JF. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol. 1993;165(4):609–631. - PubMed
-
- Allen JF. Redox control of gene expression and the function of chloroplast genomes: An hypothesis. Photosynth Res. 1993;36(2):95–102. - PubMed
-
- Allen JF, Raven JA. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J Mol Evol. 1996;42(5):482–492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

