Disease Resistance Gene Analogs (RGAs) in Plants
- PMID: 26287177
- PMCID: PMC4581296
- DOI: 10.3390/ijms160819248
Disease Resistance Gene Analogs (RGAs) in Plants
Abstract
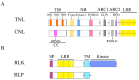
Plants have developed effective mechanisms to recognize and respond to infections caused by pathogens. Plant resistance gene analogs (RGAs), as resistance (R) gene candidates, have conserved domains and motifs that play specific roles in pathogens' resistance. Well-known RGAs are nucleotide binding site leucine rich repeats, receptor like kinases, and receptor like proteins. Others include pentatricopeptide repeats and apoplastic peroxidases. RGAs can be detected using bioinformatics tools based on their conserved structural features. Thousands of RGAs have been identified from sequenced plant genomes. High-density genome-wide RGA genetic maps are useful for designing diagnostic markers and identifying quantitative trait loci (QTL) or markers associated with plant disease resistance. This review focuses on recent advances in structures and mechanisms of RGAs, and their identification from sequenced genomes using bioinformatics tools. Applications in enhancing fine mapping and cloning of plant disease resistance genes are also discussed.
Keywords: disease resistance gene; gene mining; nucleotide binding site leucine rich repeat (NBS-LRR); pentatricopeptide repeats (PPRs); receptor like kinase (RLK); receptor like protein (RLP); resistance gene analog (RGA); small RNA (sRNA).
Figures



References
-
- Buerstmayr H., Ban T., Anderson J.A. QTL mapping and marker-assisted selection for fusarium head blight resistance in wheat: A review. Plant Breed. 2009;128:1–26. doi: 10.1111/j.1439-0523.2008.01550.x. - DOI
-
- Doane J.F., Olfert O., Mukerji M.K. Extraction precision of sieving and brine floatation for removal of wheat midge, Sitodiplosis mosellana (Diptera: Cecidomyiidae), cocoons and larvae from soil. J. Econ. Entomol. 1986;80:268–271. doi: 10.1093/jee/80.1.268. - DOI
-
- Hacquard S., Kracher B., Maekawa T., Vernaldi S., Schulze-Lefert P., Ver Loren van Themaat E. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc. Natl. Acad. Sci. USA. 2013;110:E2219–E2228. doi: 10.1073/pnas.1306807110. - DOI - PMC - PubMed
-
- Hooker A.L. The genetics and expression of resistance in plants to rusts of the genus Puccinia. Annu. Rev. Phytopathol. 1967;5:163–182. doi: 10.1146/annurev.py.05.090167.001115. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

