Intestinal Microbiota and Celiac Disease: Cause, Consequence or Co-Evolution?
- PMID: 26287240
- PMCID: PMC4555153
- DOI: 10.3390/nu7085314
Intestinal Microbiota and Celiac Disease: Cause, Consequence or Co-Evolution?
Abstract
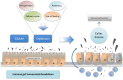
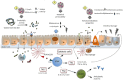
It is widely recognized that the intestinal microbiota plays a role in the initiation and perpetuation of intestinal inflammation in numerous chronic conditions. Most studies report intestinal dysbiosis in celiac disease (CD) patients, untreated and treated with a gluten-free diet (GFD), compared to healthy controls. CD patients with gastrointestinal symptoms are also known to have a different microbiota compared to patients with dermatitis herpetiformis and controls, suggesting that the microbiota is involved in disease manifestation. Furthermore, a dysbiotic microbiota seems to be associated with persistent gastrointestinal symptoms in treated CD patients, suggesting its pathogenic implication in these particular cases. GFD per se influences gut microbiota composition, and thus constitutes an inevitable confounding factor in studies conducted in CD patients. To improve our understanding of whether intestinal dysbiosis is the cause or consequence of disease, prospective studies in healthy infants at family risk of CD are underway. These studies have revealed that the CD host genotype selects for the early colonizers of the infant's gut, which together with environmental factors (e.g., breast-feeding, antibiotics, etc.) could influence the development of oral tolerance to gluten. Indeed, some CD genes and/or their altered expression play a role in bacterial colonization and sensing. In turn, intestinal dysbiosis could promote an abnormal response to gluten or other environmental CD-promoting factors (e.g., infections) in predisposed individuals. Here, we review the current knowledge of host-microbe interactions and how host genetics/epigenetics and environmental factors shape gut microbiota and may influence disease risk. We also summarize the current knowledge about the potential mechanisms of action of the intestinal microbiota and specific components that affect CD pathogenesis.
Keywords: celiac disease; dysbiosis; gluten-free diet; microbiota.
Figures
References
-
- Van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C.N.M., Bethel G., Holmes G.K.T., et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat. Genet. 2007;39:827–829. doi: 10.1038/ng2058. - DOI - PMC - PubMed
-
- Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., Bakker S.F., Bardella M.T., Bhaw-Rosun L., Castillejo G., et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

