Autophagy and Neurodegeneration: Insights from a Cultured Cell Model of ALS
- PMID: 26287246
- PMCID: PMC4588041
- DOI: 10.3390/cells4030354
Autophagy and Neurodegeneration: Insights from a Cultured Cell Model of ALS
Abstract
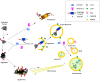
Autophagy plays a major role in the elimination of cellular waste components, the renewal of intracellular proteins and the prevention of the build-up of redundant or defective material. It is fundamental for the maintenance of homeostasis and especially important in post-mitotic neuronal cells, which, without competent autophagy, accumulate protein aggregates and degenerate. Many neurodegenerative diseases are associated with defective autophagy; however, whether altered protein turnover or accumulation of misfolded, aggregate-prone proteins is the primary insult in neurodegeneration has long been a matter of debate. Amyotrophic lateral sclerosis (ALS) is a fatal disease characterized by selective degeneration of motor neurons. Most of the ALS cases occur in sporadic forms (SALS), while 10%-15% of the cases have a positive familial history (FALS). The accumulation in the cell of misfolded/abnormal proteins is a hallmark of both SALS and FALS, and altered protein degradation due to autophagy dysregulation has been proposed to contribute to ALS pathogenesis. In this review, we focus on the main molecular features of autophagy to provide a framework for discussion of our recent findings about the role in disease pathogenesis of the ALS-linked form of the VAPB gene product, a mutant protein that drives the generation of unusual cytoplasmic inclusions.
Keywords: ALS; UPS; VAPB; aggregates; autophagy; autophagy receptors; cytoplasmic inclusions; neurodegeneration; protein degradation; proteostasis.
Figures
Similar articles
-
The ALS-linked E102Q mutation in Sigma receptor-1 leads to ER stress-mediated defects in protein homeostasis and dysregulation of RNA-binding proteins.Cell Death Differ. 2017 Oct;24(10):1655-1671. doi: 10.1038/cdd.2017.88. Epub 2017 Jun 16. Cell Death Differ. 2017. PMID: 28622300 Free PMC article.
-
Amyotrophic Lateral Sclerosis and Autophagy: Dysfunction and Therapeutic Targeting.Cells. 2020 Nov 4;9(11):2413. doi: 10.3390/cells9112413. Cells. 2020. PMID: 33158177 Free PMC article. Review.
-
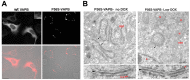
Amyotrophic lateral sclerosis-linked mutant VAPB inclusions do not interfere with protein degradation pathways or intracellular transport in a cultured cell model.PLoS One. 2014 Nov 19;9(11):e113416. doi: 10.1371/journal.pone.0113416. eCollection 2014. PLoS One. 2014. PMID: 25409455 Free PMC article.
-
ALS-associated mutant FUS inhibits macroautophagy which is restored by overexpression of Rab1.Cell Death Discov. 2015 Sep 14;1:15030. doi: 10.1038/cddiscovery.2015.30. eCollection 2015. Cell Death Discov. 2015. PMID: 27551461 Free PMC article.
-
Role of autophagy in the pathogenesis of amyotrophic lateral sclerosis.Biochim Biophys Acta. 2015 Nov;1852(11):2517-24. doi: 10.1016/j.bbadis.2015.08.005. Epub 2015 Aug 8. Biochim Biophys Acta. 2015. PMID: 26264610 Review.
Cited by
-
An open-type microdevice to improve the quality of fluorescence labeling for axonal transport analysis in neurons.Biomicrofluidics. 2019 May 9;13(3):034104. doi: 10.1063/1.5090968. eCollection 2019 May. Biomicrofluidics. 2019. PMID: 31123536 Free PMC article.
-
Repurposing carbamazepine for the treatment of amyotrophic lateral sclerosis in SOD1-G93A mouse model.CNS Neurosci Ther. 2018 Dec;24(12):1163-1174. doi: 10.1111/cns.12855. Epub 2018 Apr 14. CNS Neurosci Ther. 2018. PMID: 29656576 Free PMC article.
-
Autophagy in Stem Cell Biology: A Perspective on Stem Cell Self-Renewal and Differentiation.Stem Cells Int. 2018 Jan 21;2018:9131397. doi: 10.1155/2018/9131397. eCollection 2018. Stem Cells Int. 2018. PMID: 29765428 Free PMC article. Review.
-
SNAPIN is critical for lysosomal acidification and autophagosome maturation in macrophages.Autophagy. 2017 Feb;13(2):285-301. doi: 10.1080/15548627.2016.1261238. Epub 2016 Dec 8. Autophagy. 2017. PMID: 27929705 Free PMC article.
-
Rbfox-Splicing Factors Maintain Skeletal Muscle Mass by Regulating Calpain3 and Proteostasis.Cell Rep. 2018 Jul 3;24(1):197-208. doi: 10.1016/j.celrep.2018.06.017. Cell Rep. 2018. PMID: 29972780 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

