Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6
- PMID: 26287468
- PMCID: PMC4697747
- DOI: 10.1038/nature15252
Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6
Abstract
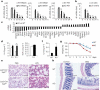
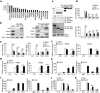
Epigenetic modifiers have fundamental roles in defining unique cellular identity through the establishment and maintenance of lineage-specific chromatin and methylation status. Several DNA modifications such as 5-hydroxymethylcytosine (5hmC) are catalysed by the ten eleven translocation (Tet) methylcytosine dioxygenase family members, and the roles of Tet proteins in regulating chromatin architecture and gene transcription independently of DNA methylation have been gradually uncovered. However, the regulation of immunity and inflammation by Tet proteins independent of their role in modulating DNA methylation remains largely unknown. Here we show that Tet2 selectively mediates active repression of interleukin-6 (IL-6) transcription during inflammation resolution in innate myeloid cells, including dendritic cells and macrophages. Loss of Tet2 resulted in the upregulation of several inflammatory mediators, including IL-6, at late phase during the response to lipopolysaccharide challenge. Tet2-deficient mice were more susceptible to endotoxin shock and dextran-sulfate-sodium-induced colitis, displaying a more severe inflammatory phenotype and increased IL-6 production compared to wild-type mice. IκBζ, an IL-6-specific transcription factor, mediated specific targeting of Tet2 to the Il6 promoter, further indicating opposite regulatory roles of IκBζ at initial and resolution phases of inflammation. For the repression mechanism, independent of DNA methylation and hydroxymethylation, Tet2 recruited Hdac2 and repressed transcription of Il6 via histone deacetylation. We provide mechanistic evidence for the gene-specific transcription repression activity of Tet2 via histone deacetylation and for the prevention of constant transcription activation at the chromatin level for resolving inflammation.
Figures
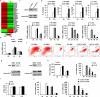

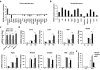
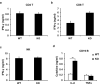


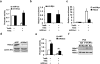


Comment in
-
Inflammation: TET2: the terminator.Nat Rev Immunol. 2015 Oct;15(10):598. doi: 10.1038/nri3912. Epub 2015 Sep 11. Nat Rev Immunol. 2015. PMID: 26358395 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

