Identifying new targets in leukemogenesis using computational approaches
- PMID: 26288567
- PMCID: PMC4537869
- DOI: 10.1016/j.sjbs.2015.01.012
Identifying new targets in leukemogenesis using computational approaches
Abstract
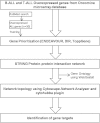
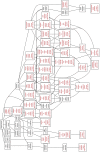
There is a need to identify novel targets in Acute Lymphoblastic Leukemia (ALL), a hematopoietic cancer affecting children, to improve our understanding of disease biology and that can be used for developing new therapeutics. Hence, the aim of our study was to find new genes as targets using in silico studies; for this we retrieved the top 10% overexpressed genes from Oncomine public domain microarray expression database; 530 overexpressed genes were short-listed from Oncomine database. Then, using prioritization tools such as ENDEAVOUR, DIR and TOPPGene online tools, we found fifty-four genes common to the three prioritization tools which formed our candidate leukemogenic genes for this study. As per the protocol we selected thirty training genes from PubMed. The prioritized and training genes were then used to construct STRING functional association network, which was further analyzed using cytoHubba hub analysis tool to investigate new genes which could form drug targets in leukemia. Analysis of the STRING protein network built from these prioritized and training genes led to identification of two hub genes, SMAD2 and CDK9, which were not implicated in leukemogenesis earlier. Filtering out from several hundred genes in the network we also found MEN1, HDAC1 and LCK genes, which re-emphasized the important role of these genes in leukemogenesis. This is the first report on these five additional signature genes in leukemogenesis. We propose these as new targets for developing novel therapeutics and also as biomarkers in leukemogenesis, which could be important for prognosis and diagnosis.
Keywords: Acute Lymphoblastic Leukemia (ALL); Gene prioritization; Microarray analysis; Protein interaction network; Therapeutic targets.
Figures


Similar articles
-
Identifying Key Lysosome-Related Genes Associated with Drug-Resistant Breast Cancer Using Computational and Systems Biology Approach.Iran J Pharm Res. 2022 Oct 15;21(1):e130342. doi: 10.5812/ijpr-130342. eCollection 2022 Dec. Iran J Pharm Res. 2022. PMID: 36915401 Free PMC article.
-
In silico evidence of signaling pathways of notch mediated networks in leukemia.Comput Struct Biotechnol J. 2012 Nov 19;1:e201207005. doi: 10.5936/csbj.201207005. eCollection 2012. Comput Struct Biotechnol J. 2012. PMID: 24688641 Free PMC article.
-
Identification of key biomarkers associated with development and prognosis in patients with ovarian carcinoma: evidence from bioinformatic analysis.J Ovarian Res. 2019 Nov 15;12(1):110. doi: 10.1186/s13048-019-0578-1. J Ovarian Res. 2019. PMID: 31729978 Free PMC article.
-
Genomic assessment of pediatric acute leukemia.Cancer J. 2005 Jul-Aug;11(4):268-82. doi: 10.1097/00130404-200507000-00003. Cancer J. 2005. PMID: 16197717 Review.
-
Gene Prioritization and Network Topology Analysis of Targeted Genes for Acquired Taxane Resistance by Meta-Analysis.Crit Rev Eukaryot Gene Expr. 2019;29(6):581-597. doi: 10.1615/CritRevEukaryotGeneExpr.2019026317. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422012 Review.
Cited by
-
Copy number variant analysis for syndromic congenital heart disease in the Chinese population.Hum Genomics. 2022 Oct 31;16(1):51. doi: 10.1186/s40246-022-00426-8. Hum Genomics. 2022. PMID: 36316717 Free PMC article.
-
A prognostic nomogram for lung adenocarcinoma based on immune-infiltrating Treg-related genes: from bench to bedside.Transl Lung Cancer Res. 2021 Jan;10(1):167-182. doi: 10.21037/tlcr-20-822. Transl Lung Cancer Res. 2021. PMID: 33569302 Free PMC article.
-
CDK9 Inhibitor Induces the Apoptosis of B-Cell Acute Lymphocytic Leukemia by Inhibiting c-Myc-Mediated Glycolytic Metabolism.Front Cell Dev Biol. 2021 Mar 4;9:641271. doi: 10.3389/fcell.2021.641271. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33748130 Free PMC article.
-
Methylation-Driven Gene PLAU as a Potential Prognostic Marker for Differential Thyroid Carcinoma.Front Cell Dev Biol. 2022 Jan 24;10:819484. doi: 10.3389/fcell.2022.819484. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35141223 Free PMC article.
References
-
- Andersson A., Ritz C., Lindgren D., Edén P., Lassen C., Heldrup J., Olofsson T., Råde J., Fontes M., Porwit-Macdonald A., Behrendtz M., Höglund M., Johansson B., Fioretos T. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. - PubMed
-
- Arai F., Suda T. Maintenance of quiescent hematopoietic stem cells in the osteoblastic niche. Ann. N. Y. Acad. Sci. 2007;1106:41–53. - PubMed
-
- Aref S., Mabed M., El-Sherbiny M., Selim T., Metwaly A. Cyclin D1 expression in acute leukemia. Hematology. 2006;11:31–34. - PubMed
-
- Assenov Y., Ramírez F., Schelhorn S.E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

