Structural mechanism for signal transduction in RXR nuclear receptor heterodimers
- PMID: 26289479
- PMCID: PMC4547401
- DOI: 10.1038/ncomms9013
Structural mechanism for signal transduction in RXR nuclear receptor heterodimers
Abstract
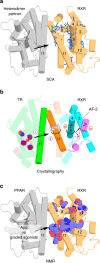
A subset of nuclear receptors (NRs) function as obligate heterodimers with retinoid X receptor (RXR), allowing integration of ligand-dependent signals across the dimer interface via an unknown structural mechanism. Using nuclear magnetic resonance (NMR) spectroscopy, x-ray crystallography and hydrogen/deuterium exchange (HDX) mass spectrometry, here we show an allosteric mechanism through which RXR co-operates with a permissive dimer partner, peroxisome proliferator-activated receptor (PPAR)-γ, while rendered generally unresponsive by a non-permissive dimer partner, thyroid hormone (TR) receptor. Amino acid residues that mediate this allosteric mechanism comprise an evolutionarily conserved network discovered by statistical coupling analysis (SCA). This SCA network acts as a signalling rheostat to integrate signals between dimer partners, ligands and coregulator-binding sites, thereby affecting signal transmission in RXR heterodimers. These findings define rules guiding how NRs integrate two ligand-dependent signalling pathways into RXR heterodimer-specific responses.
Conflict of interest statement
Kendall Nettles is a consultant for Genentech.
Figures



References
-
- Shulman A. I. & Mangelsdorf D. J. Retinoid x receptor heterodimers in the metabolic syndrome. N. Engl. J. Med. 353, 604–615 (2005). - PubMed
-
- Rosenfeld M. G., Lunyak V. V. & Glass C. K. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20, 1405–1428 (2006). - PubMed
-
- Shang Y. & Brown M. Molecular determinants for the tissue specificity of SERMs. Science 295, 2465–2468 (2002). - PubMed
-
- Marciano D. P. et al. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARgamma, RORs, and Rev-erbs. Cell Metab. 19, 193–208 (2014). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 DK105825/DK/NIDDK NIH HHS/United States
- MH084512/MH/NIMH NIH HHS/United States
- RR019077/RR/NCRR NIH HHS/United States
- R00 DK103116/DK/NIDDK NIH HHS/United States
- RR027270/RR/NCRR NIH HHS/United States
- S10 RR027755/RR/NCRR NIH HHS/United States
- RR027755/RR/NCRR NIH HHS/United States
- GM114420/GM/NIGMS NIH HHS/United States
- GM063855/GM/NIGMS NIH HHS/United States
- R01 GM084041/GM/NIGMS NIH HHS/United States
- S10 RR027270/RR/NCRR NIH HHS/United States
- R01 GM063855/GM/NIGMS NIH HHS/United States
- CA132022/CA/NCI NIH HHS/United States
- R01 DK101871/DK/NIDDK NIH HHS/United States
- U54 MH084512/MH/NIMH NIH HHS/United States
- F32 DK097890/DK/NIDDK NIH HHS/United States
- DK103116/DK/NIDDK NIH HHS/United States
- DK097890/DK/NIDDK NIH HHS/United States
- GM084041/GM/NIGMS NIH HHS/United States
- S10 RR019077/RR/NCRR NIH HHS/United States
- R01 GM114420/GM/NIGMS NIH HHS/United States
- K99 DK103116/DK/NIDDK NIH HHS/United States
- R33 CA132022/CA/NCI NIH HHS/United States
- DK101871/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

