Quantitative analysis of recombination between YFP and CFP genes of FRET biosensors introduced by lentiviral or retroviral gene transfer
- PMID: 26290434
- PMCID: PMC4542544
- DOI: 10.1038/srep13283
Quantitative analysis of recombination between YFP and CFP genes of FRET biosensors introduced by lentiviral or retroviral gene transfer
Abstract
Biosensors based on the principle of Förster (or fluorescence) resonance energy transfer (FRET) have been developed to visualize spatio-temporal dynamics of signalling molecules in living cells. Many of them adopt a backbone of intramolecular FRET biosensor with a cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) as donor and acceptor, respectively. However, there remains the difficulty of establishing cells stably expressing FRET biosensors with a YFP and CFP pair by lentiviral or retroviral gene transfer, due to the high incidence of recombination between YFP and CFP genes. To address this, we examined the effects of codon-diversification of YFP on the recombination of FRET biosensors introduced by lentivirus or retrovirus. The YFP gene that was fully codon-optimized to E.coli evaded the recombination in lentiviral or retroviral gene transfer, but the partially codon-diversified YFP did not. Further, the length of spacer between YFP and CFP genes clearly affected recombination efficiency, suggesting that the intramolecular template switching occurred in the reverse-transcription process. The simple mathematical model reproduced the experimental data sufficiently, yielding a recombination rate of 0.002-0.005 per base. Together, these results show that the codon-diversified YFP is a useful tool for expressing FRET biosensors by lentiviral or retroviral gene transfer.
Figures
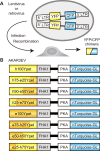

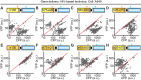
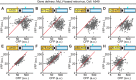

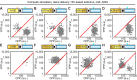
Similar articles
-
Booster, a Red-Shifted Genetically Encoded Förster Resonance Energy Transfer (FRET) Biosensor Compatible with Cyan Fluorescent Protein/Yellow Fluorescent Protein-Based FRET Biosensors and Blue Light-Responsive Optogenetic Tools.ACS Sens. 2020 Mar 27;5(3):719-730. doi: 10.1021/acssensors.9b01941. Epub 2020 Feb 26. ACS Sens. 2020. PMID: 32101394
-
A flow cytometric method to detect protein-protein interaction in living cells by directly visualizing donor fluorophore quenching during CFP-->YFP fluorescence resonance energy transfer (FRET).Cytometry A. 2003 Oct;55(2):71-85. doi: 10.1002/cyto.a.10073. Cytometry A. 2003. PMID: 14505312
-
Flow cytometric measurement of fluorescence (Förster) resonance energy transfer from cyan fluorescent protein to yellow fluorescent protein using single-laser excitation at 458 nm.Cytometry A. 2003 May;53(1):39-54. doi: 10.1002/cyto.a.10037. Cytometry A. 2003. PMID: 12701131
-
Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics.Biotechnol Lett. 2006 Dec;28(24):1971-82. doi: 10.1007/s10529-006-9193-5. Epub 2006 Oct 5. Biotechnol Lett. 2006. PMID: 17021660 Review.
-
Fluorescence resonance energy transfer imaging of cell signaling from in vitro to in vivo: basis of biosensor construction, live imaging, and image processing.Dev Growth Differ. 2013 May;55(4):515-22. doi: 10.1111/dgd.12039. Epub 2013 Feb 7. Dev Growth Differ. 2013. PMID: 23387795 Review.
Cited by
-
What do we know about dynamic glucose-enhanced (DGE) MRI and how close is it to the clinics? Horizon 2020 GLINT consortium report.MAGMA. 2022 Feb;35(1):87-104. doi: 10.1007/s10334-021-00994-1. Epub 2022 Jan 15. MAGMA. 2022. PMID: 35032288 Free PMC article. Review.
-
Single-Cell Imaging of ERK Signaling Using Fluorescent Biosensors.Methods Mol Biol. 2017;1636:35-59. doi: 10.1007/978-1-4939-7154-1_3. Methods Mol Biol. 2017. PMID: 28730471 Free PMC article.
-
Abundance-biased codon diversification prevents recombination in AAV production and ensures robust in vivo expression of functional FRET sensors.Commun Biol. 2025 Aug 19;8(1):1244. doi: 10.1038/s42003-025-08677-6. Commun Biol. 2025. PMID: 40830588 Free PMC article.
-
Live-Cell FRET Imaging Reveals a Role of Extracellular Signal-Regulated Kinase Activity Dynamics in Thymocyte Motility.iScience. 2018 Dec 21;10:98-113. doi: 10.1016/j.isci.2018.11.025. Epub 2018 Nov 20. iScience. 2018. PMID: 30508722 Free PMC article.
-
Shedding new light on RhoA signalling as a drug target in vivo using a novel RhoA-FRET biosensor mouse.Small GTPases. 2020 Jul;11(4):240-247. doi: 10.1080/21541248.2018.1438024. Epub 2018 Mar 21. Small GTPases. 2020. PMID: 29457531 Free PMC article.
References
-
- Miyawaki A. et al. Fluorescent indicators for Ca2+based on green fluorescent proteins and calmodulin. 388, 882–887 (1997). - PubMed
-
- Sato M., Ueda Y., Takagi T. & Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 5, 1016–22 (2003). - PubMed
-
- Mochizuki N. et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature 411, 1065–8 (2001). - PubMed
-
- Aoki K. et al. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell 52, 529–40 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

